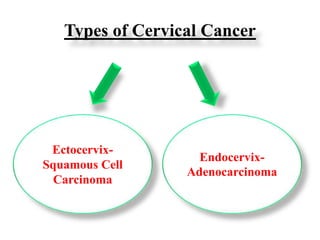
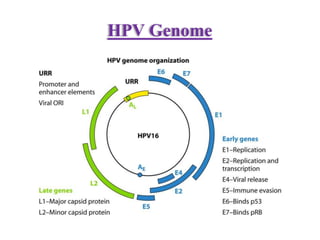
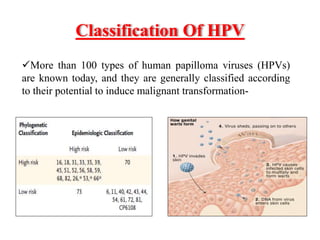
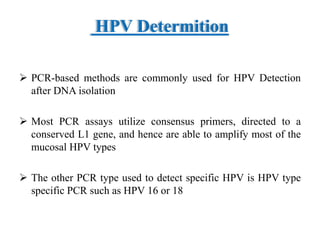
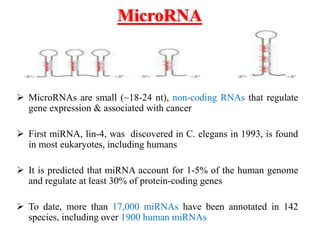
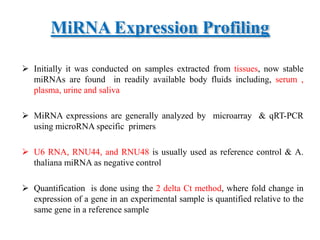
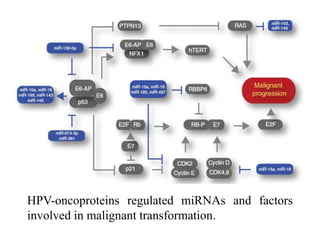
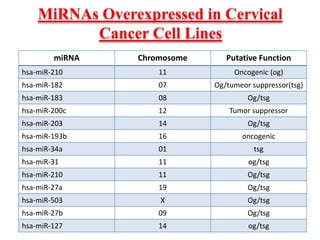
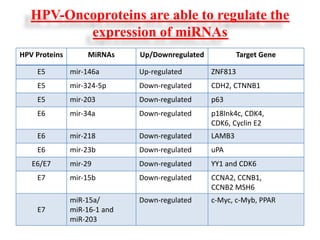
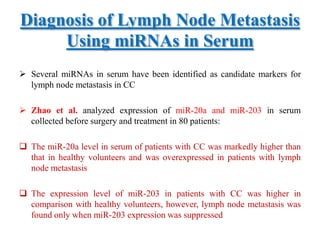
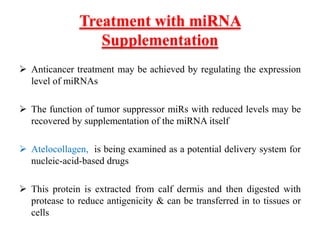
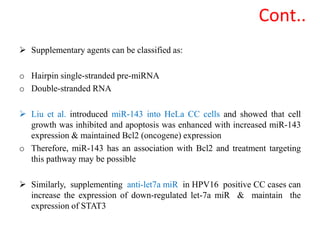
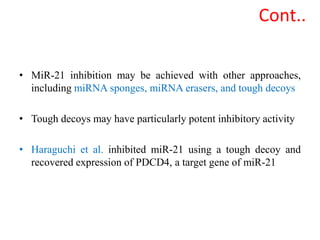
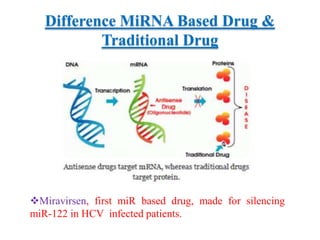
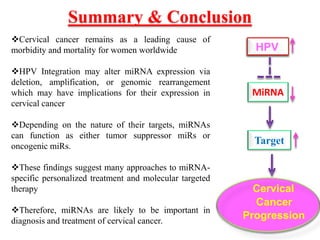
This document discusses cervical cancer, its causes, symptoms, diagnosis, and treatment. Some key points:
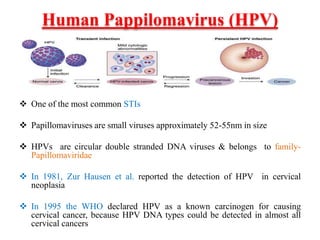
- Cervical cancer is the fourth most common cancer in women worldwide and is caused by human papillomavirus (HPV) infection in over 90% of cases.
- Other risk factors include multiple sexual partners, young age of first intercourse, smoking, and a weakened immune system.
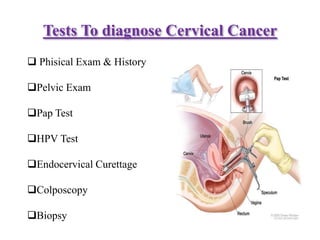
- Symptoms can include abnormal bleeding or discharge. Diagnosis involves exams, Pap tests, HPV tests, biopsies and assessing the cancer stage.
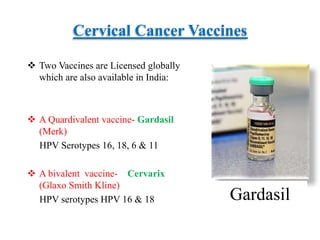
- Cervical cancer is typically treated through surgery, radiation therapy, chemotherapy, or a combination depending on the cancer stage and grade. Vaccines can