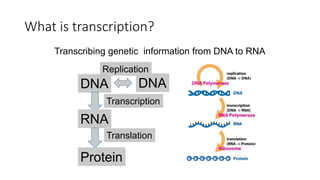
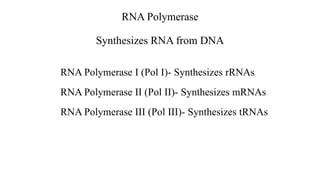
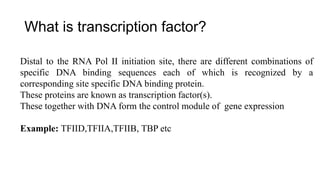
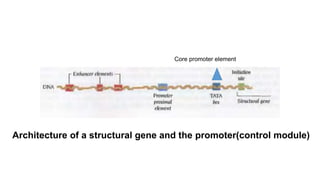
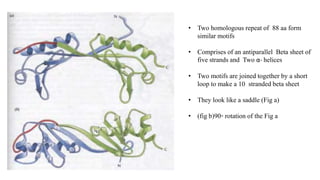
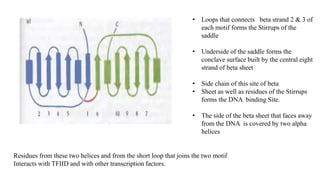
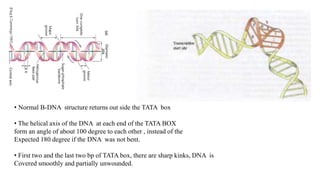
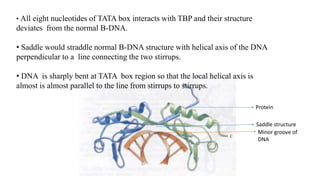
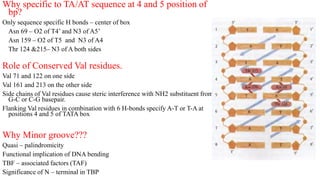
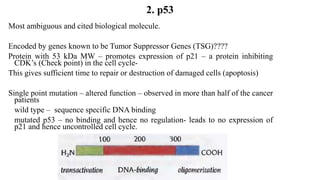
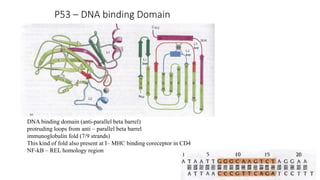
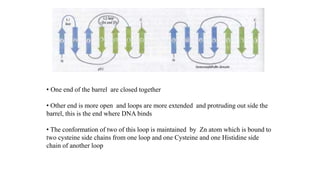
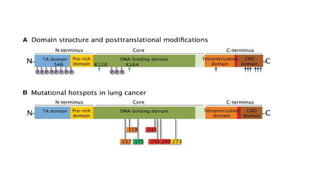
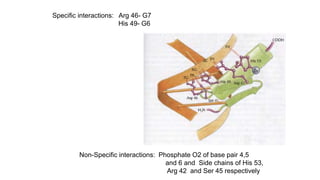
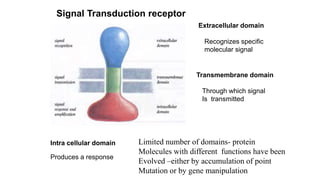
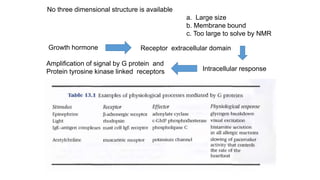
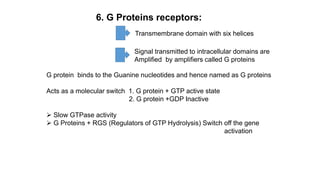
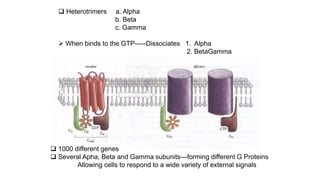
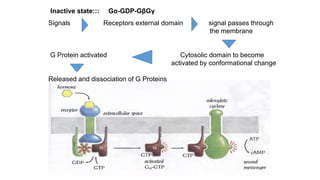
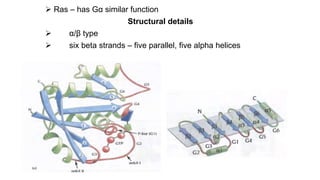
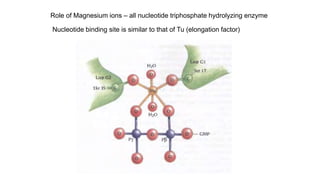
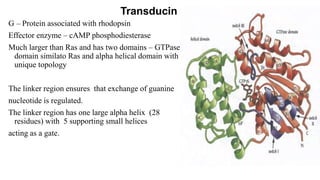
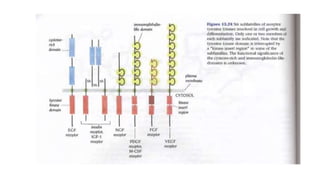
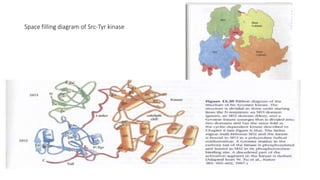
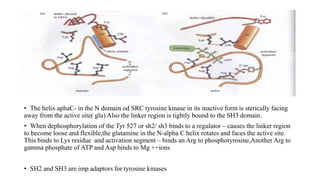
1. The document discusses the structure-function correlation in various proteins including transcription factors like TATA box binding proteins, tumor suppressor protein p53, and signal transducer proteins.
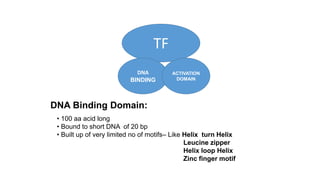
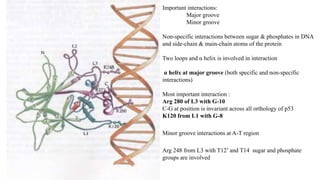
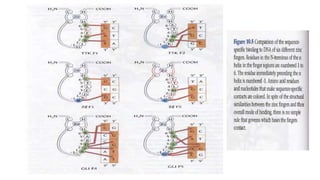
2. It describes the DNA binding domains of transcription factors like TATA box binding protein which binds to the minor groove of DNA, and p53 which uses loops and alpha helices to interact with DNA in both the major and minor grooves.
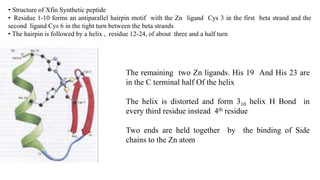
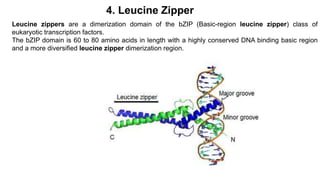
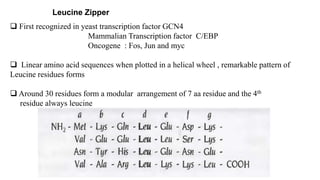
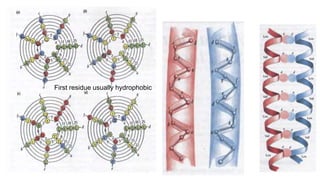
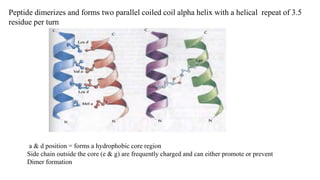
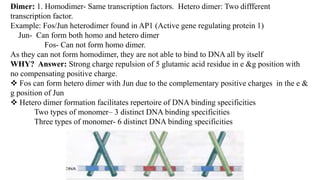
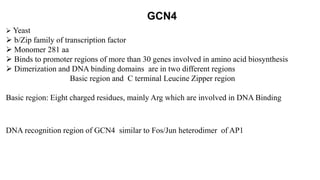
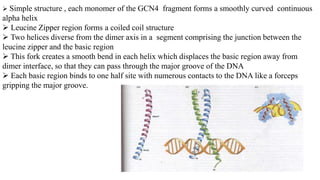
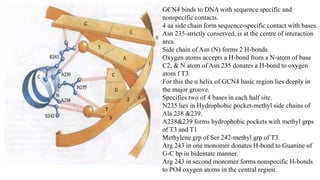
3. Zinc finger proteins and leucine zipper domains are discussed as examples of DNA binding motifs, with zinc fingers using cysteine and histidine residues to coordinate zinc ions and form DNA interacting loops and helices, while leucine zippers enable dimerization of transcription factors via