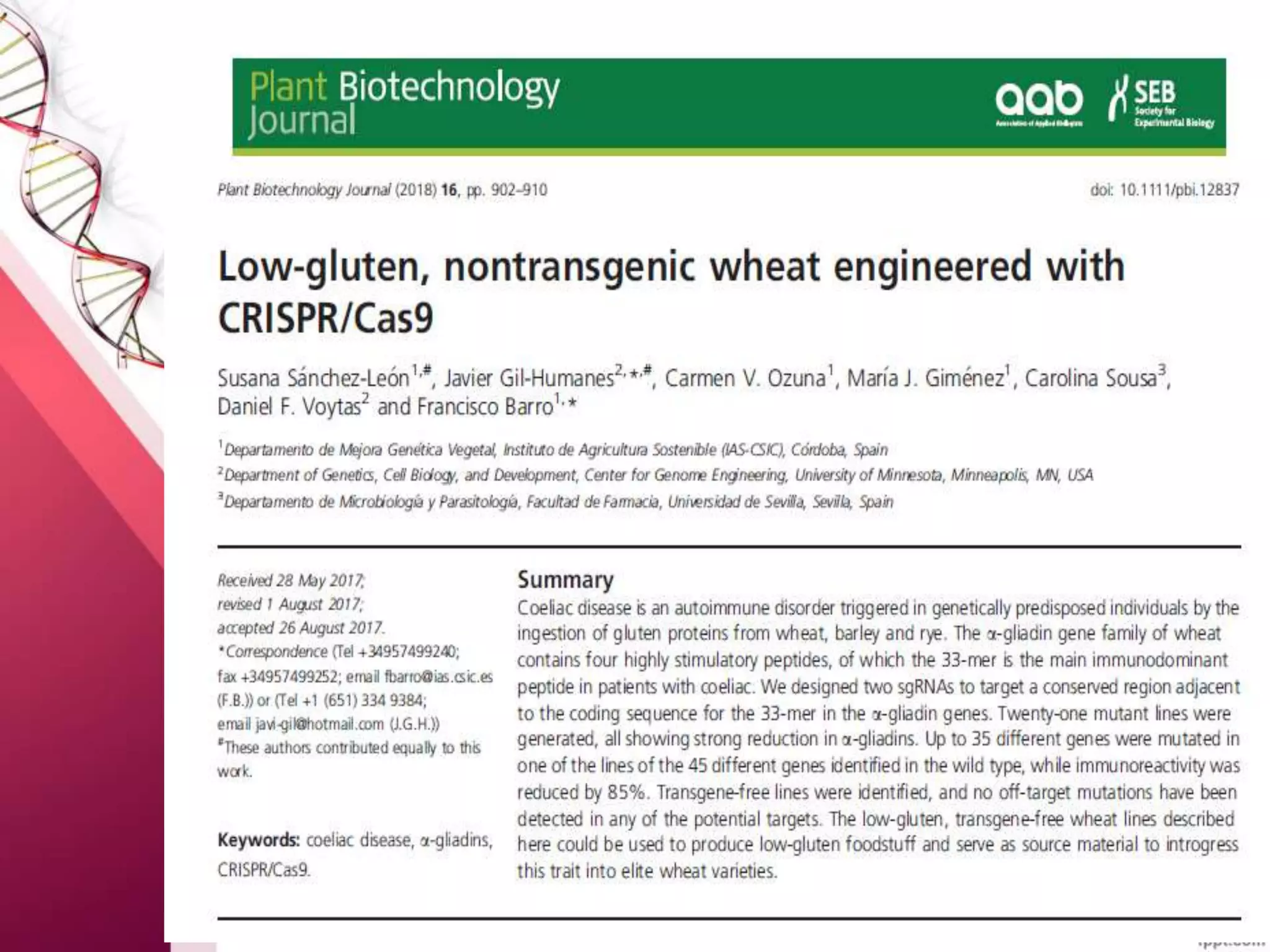
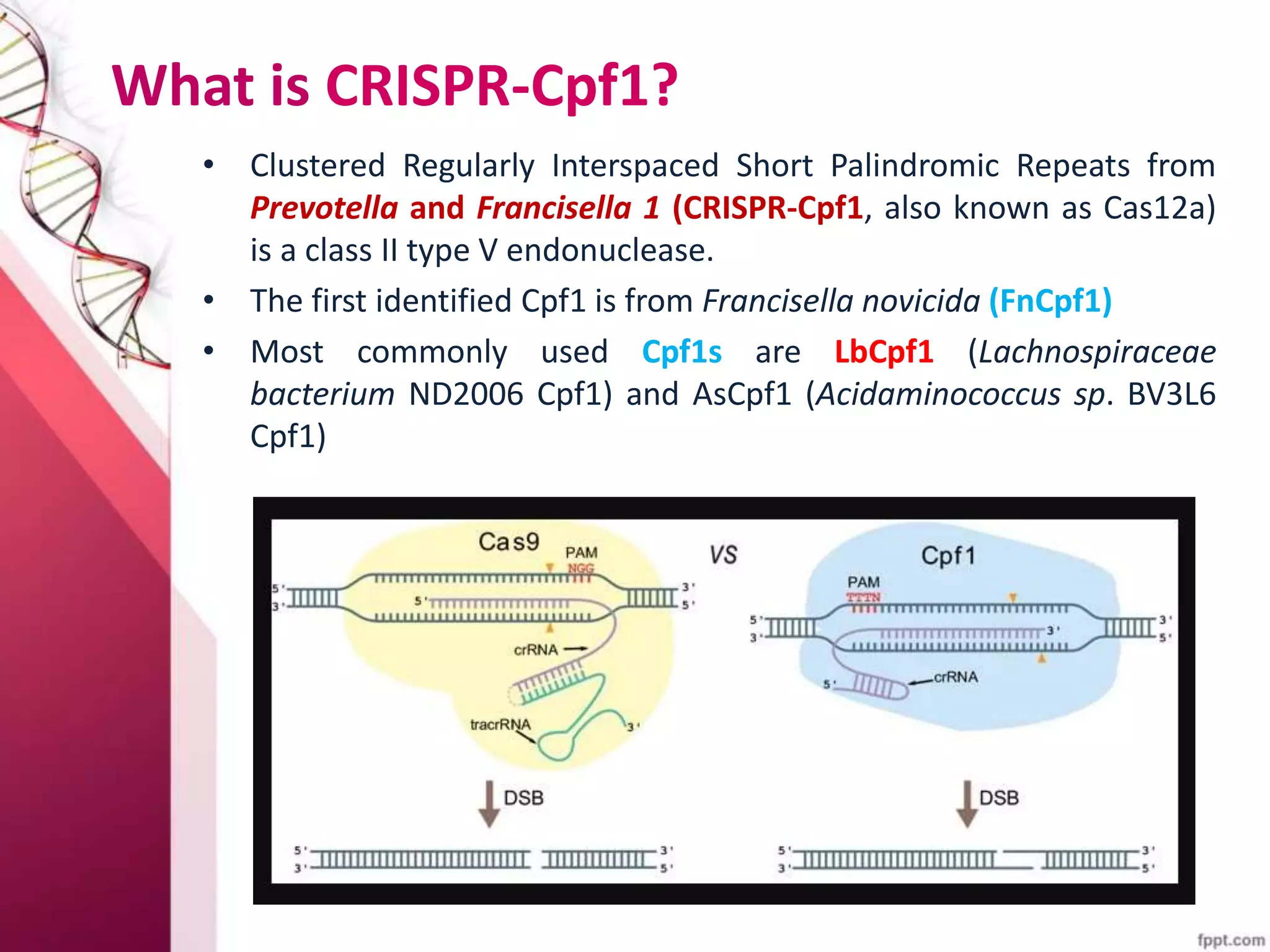
This document discusses the use of CRISPR-Cas9 genome editing in crop improvement. It begins with an introduction to CRISPR-Cas9 and its mechanism of action. It then discusses the discovery of CRISPR and key scientists involved. Several case studies on using CRISPR to edit rice genes for disease resistance and hybrid seed production are summarized. Achievements using CRISPR in rice, horticulture crops, and other field crops are briefly outlined. The document concludes that CRISPR provides a simple and efficient tool for genome editing in plants.
![CRISPR
[Clustered Regularly Interspaced Short
Palindromic Repeats]
in Crop Improvement
PARTHASARATHI.G
2018-630-812
II M.Sc(Ag)
Dept of PBG](https://image.slidesharecdn.com/crisprppt-191116194118/75/CRISPR-in-crop-Improvement-CRISPR-Cas-Genome-editing-tool-1-2048.jpg)
![Introduction
• CRISPR-Cas9 is used to edit parts of
the genome by removing, adding or altering
sections of the DNA sequence, based on
bacterial adaptive immune system .
• Key Elements
i) CRISPR sequence
[clustered regularly interspaced short
palindromic repeats]
ii) Cas9 (nucleases)](https://image.slidesharecdn.com/crisprppt-191116194118/75/CRISPR-in-crop-Improvement-CRISPR-Cas-Genome-editing-tool-3-2048.jpg)
![Mechanism of CRISPR/Cas9 complex
• Creating Nuclease induced DSBs (Double stranded
breaks) .
• Repairing DSBs in either one of the two pathways.
»NHEJ
[Non-homologous End Joining]
»HDR repair
[Homology Directed Repair]
• It is unique and flexible owing to its dependence
on RNA as the moiety that targets the nuclease to
a desired DNA sequence.](https://image.slidesharecdn.com/crisprppt-191116194118/75/CRISPR-in-crop-Improvement-CRISPR-Cas-Genome-editing-tool-4-2048.jpg)