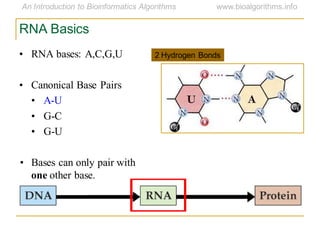
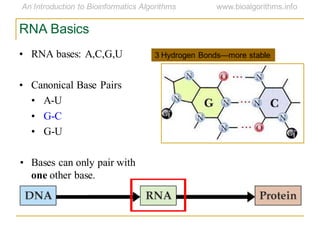
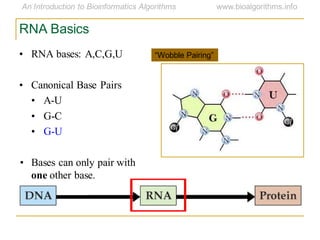
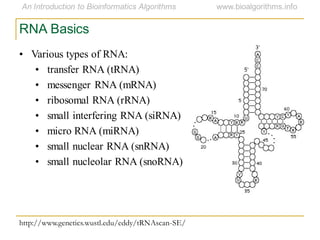
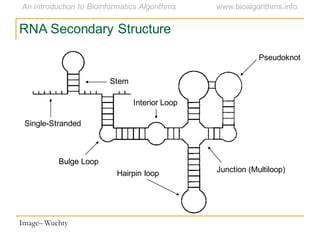
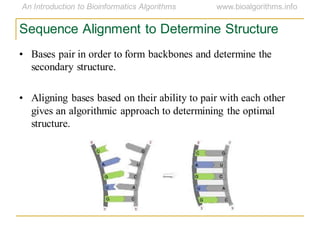
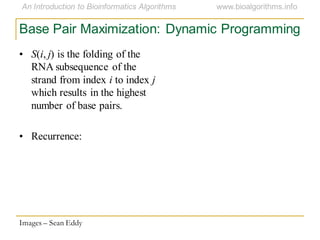
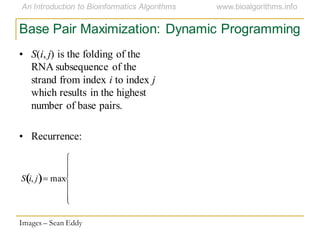
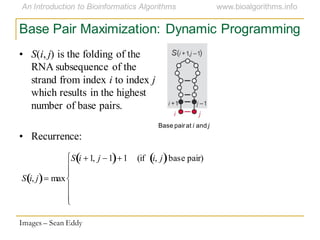
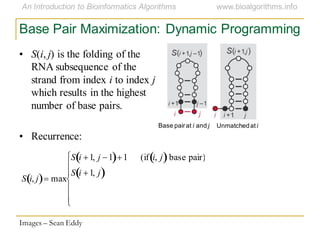
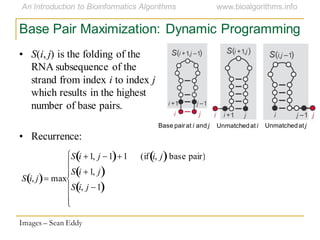
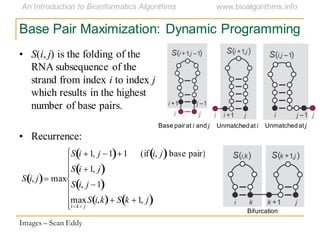
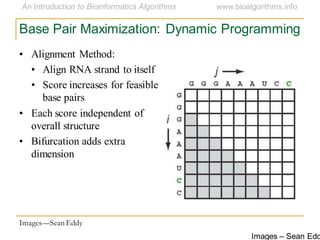
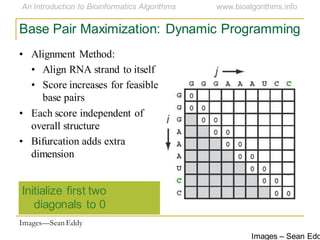
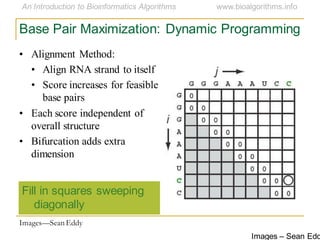
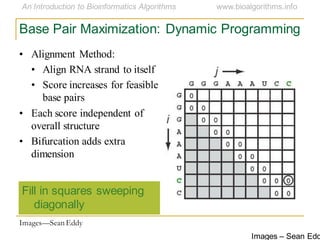
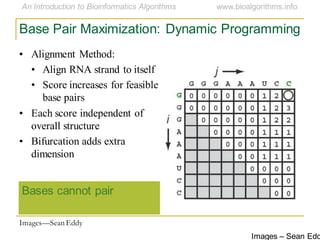
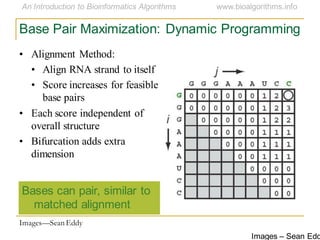
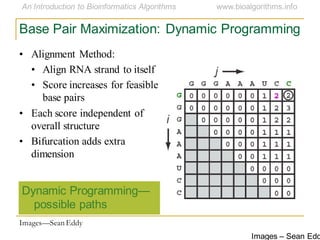
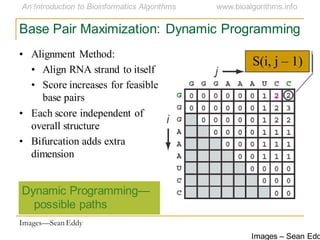
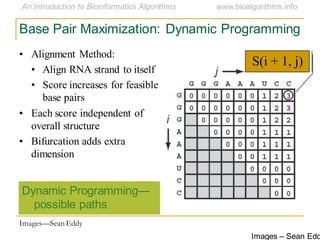
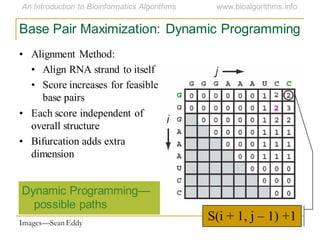
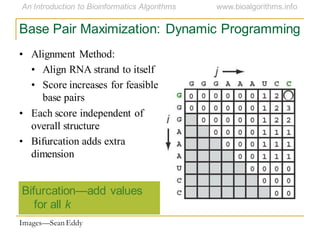
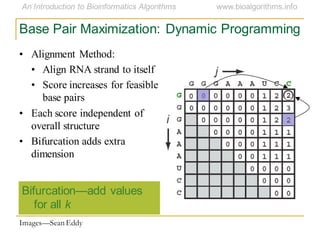
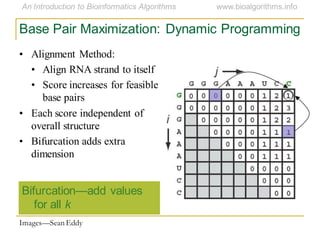
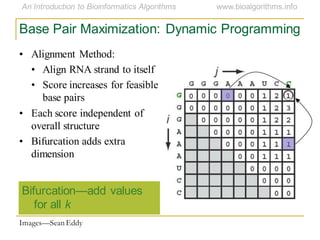
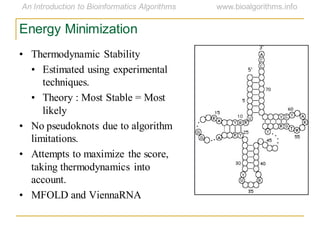
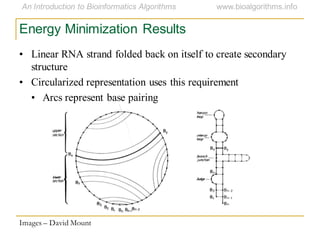
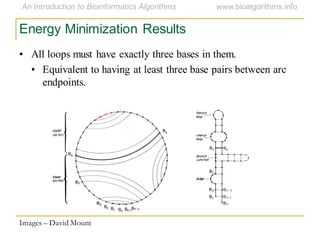
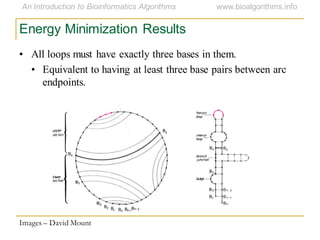
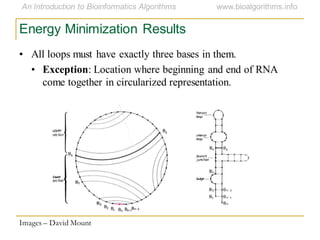
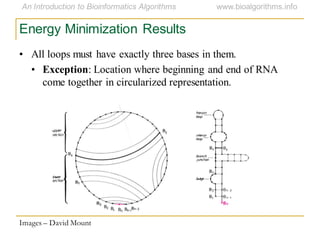
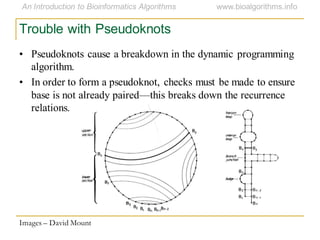
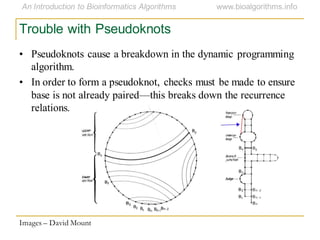
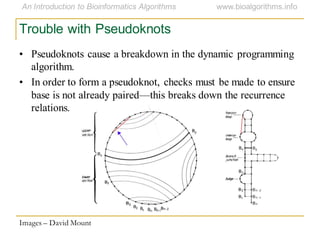
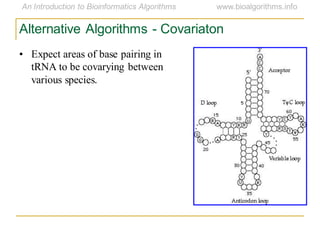
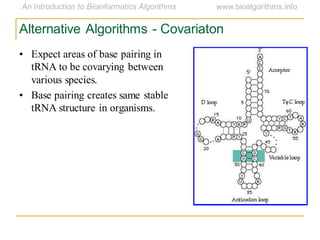
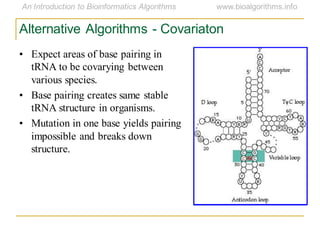
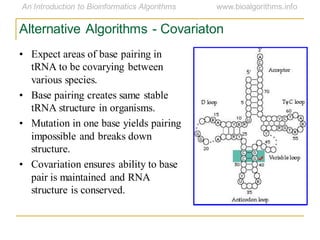
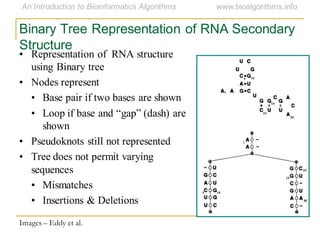
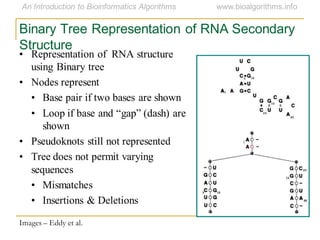
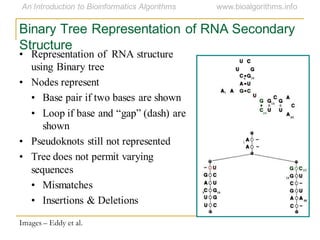
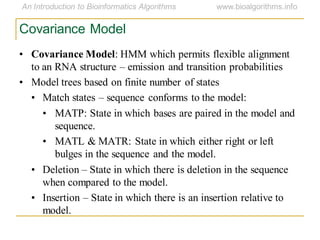
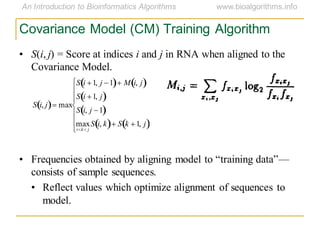
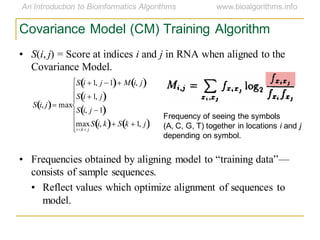
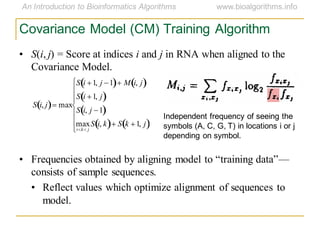
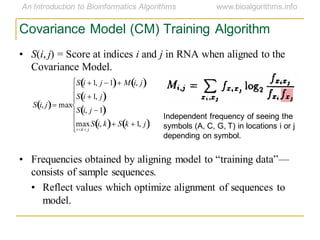
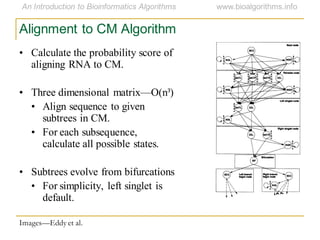
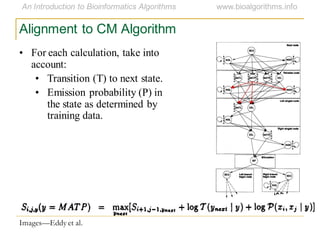
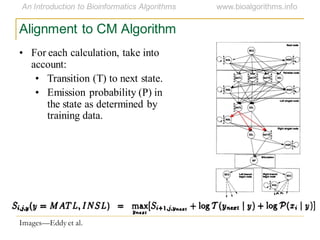
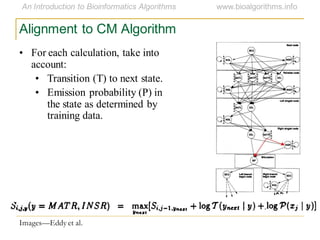
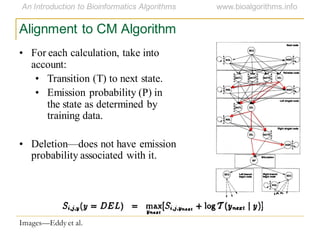
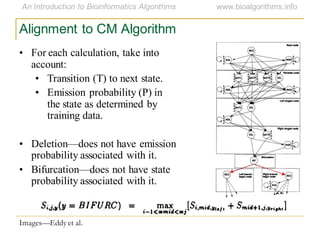
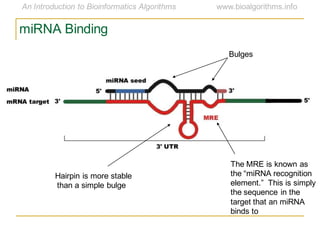
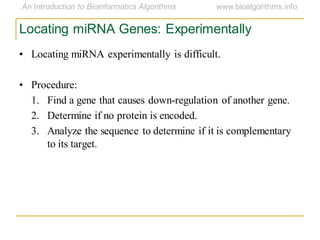
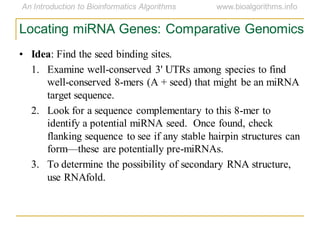
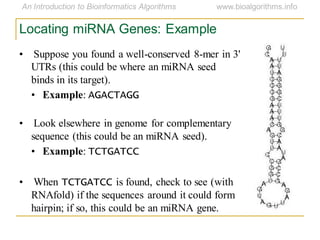
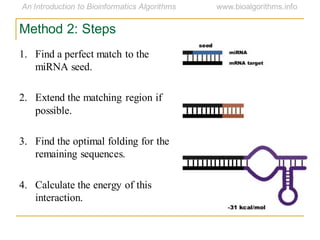
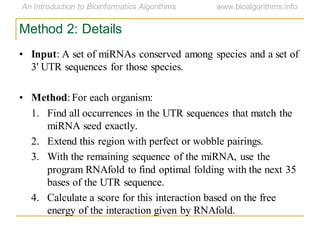
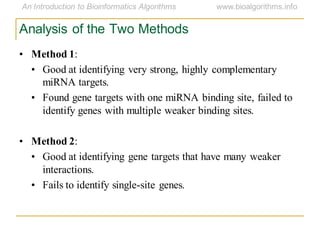
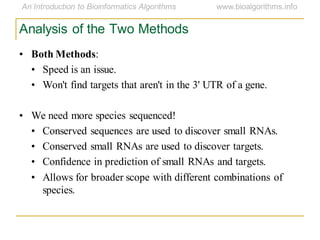
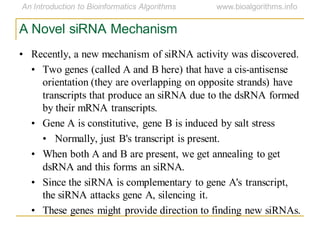
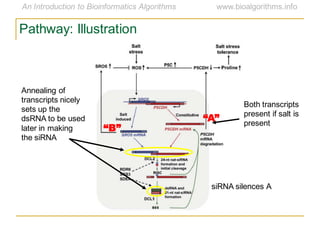
This document discusses methods for predicting and analyzing the secondary structure of RNA molecules. It begins by covering RNA folding basics like canonical base pairs. It then describes two main approaches to secondary structure prediction: dynamic programming which aims to maximize base pairing, and energy minimization which considers thermodynamic stability. Dynamic programming uses a recurrence relation and bifurcation in a dynamic programming algorithm. Energy minimization computes the single most stable structure but may not be biologically accurate. Covariance models incorporate similarity-based methods and use hidden Markov models to represent consensus structures allowing for flexible sequence alignments based on observed co-varying mutations.