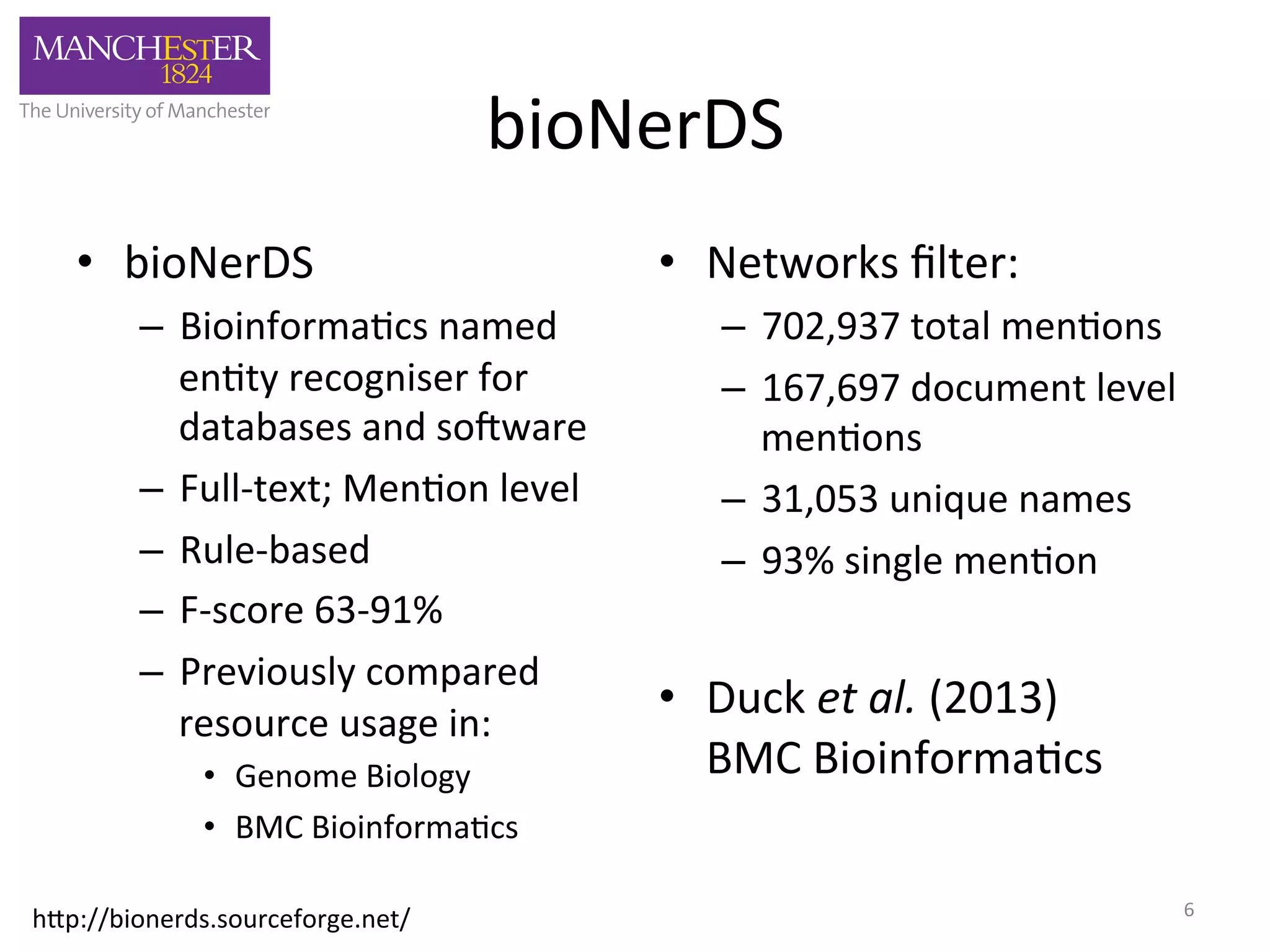
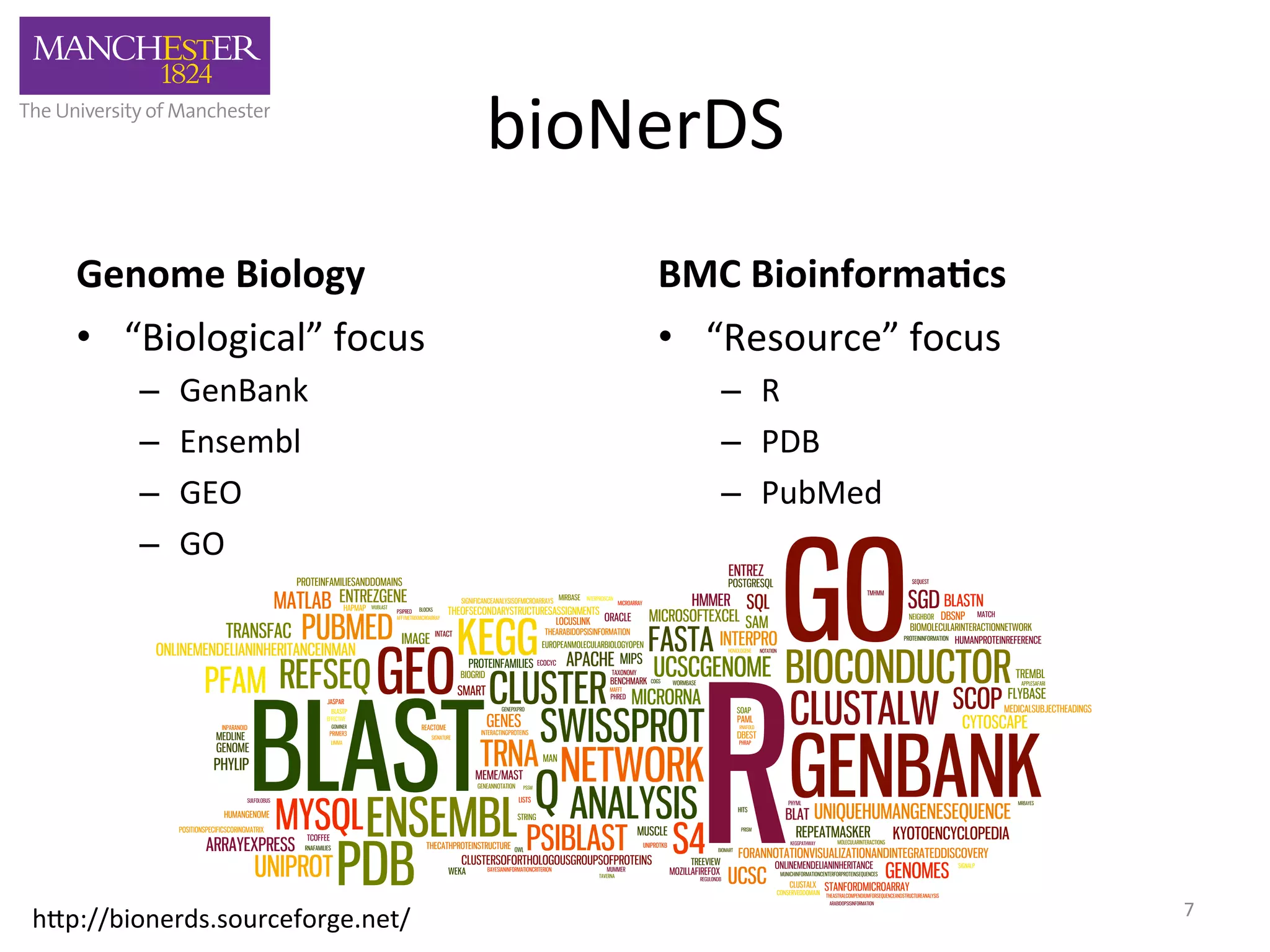
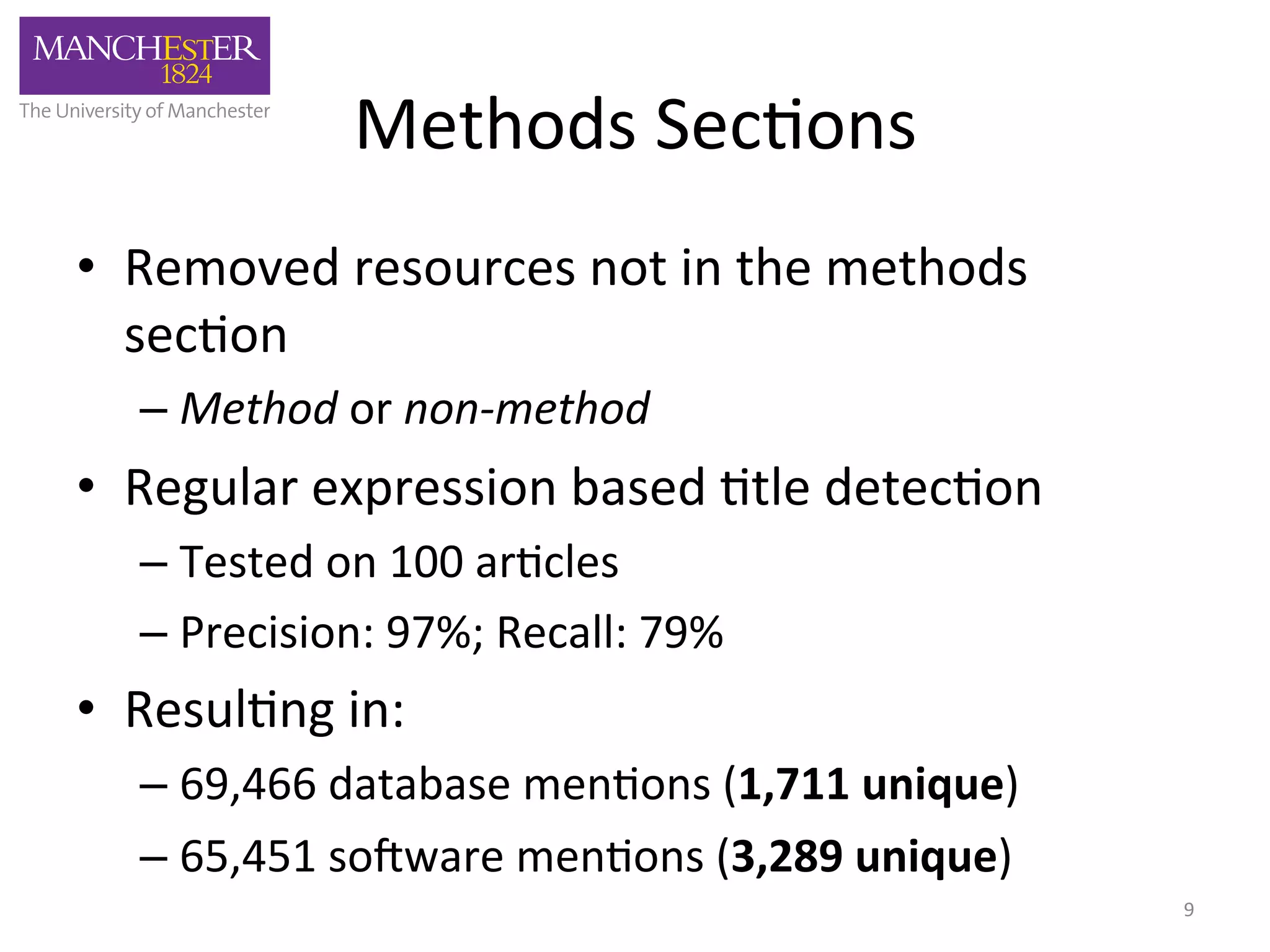
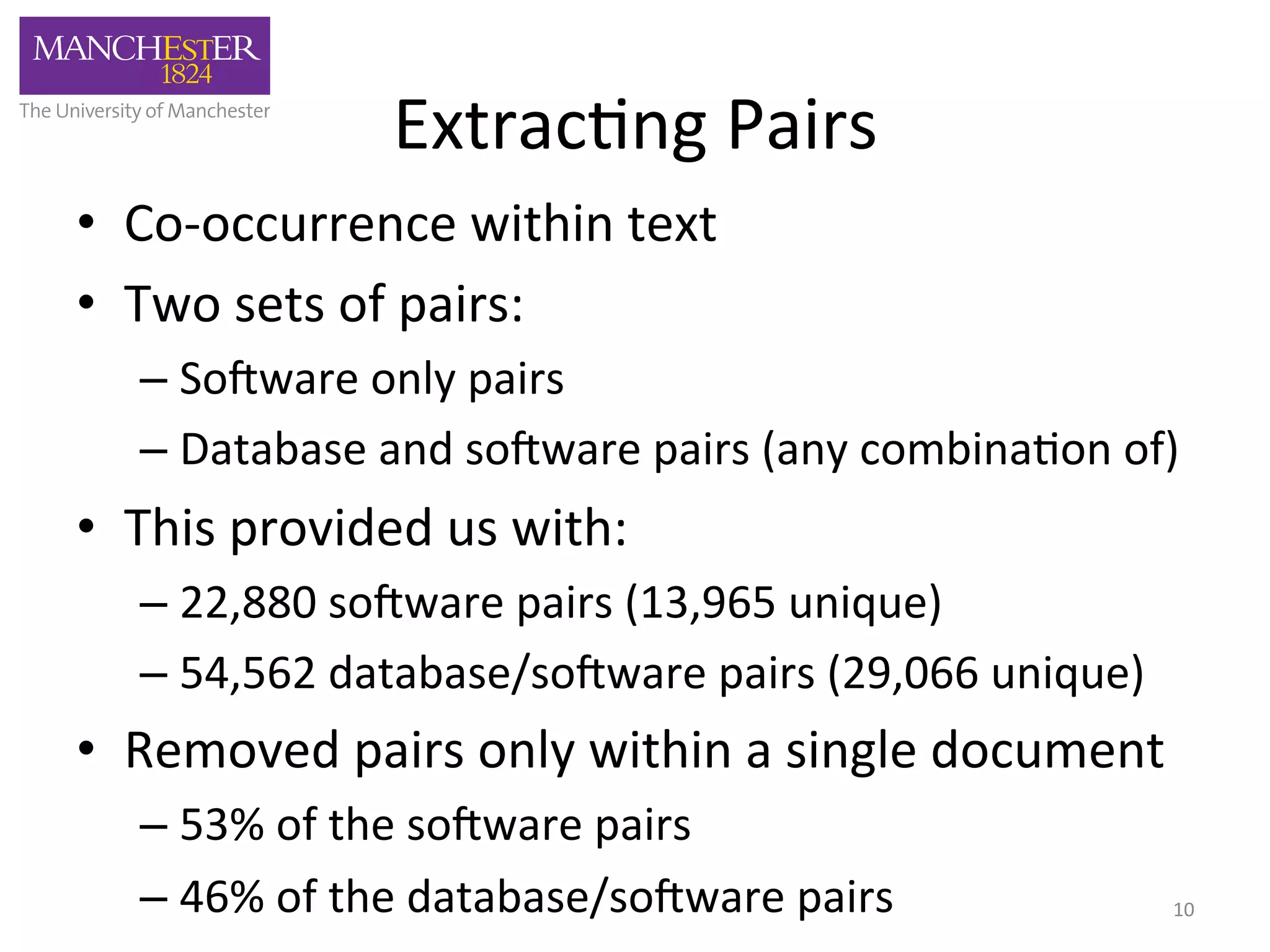
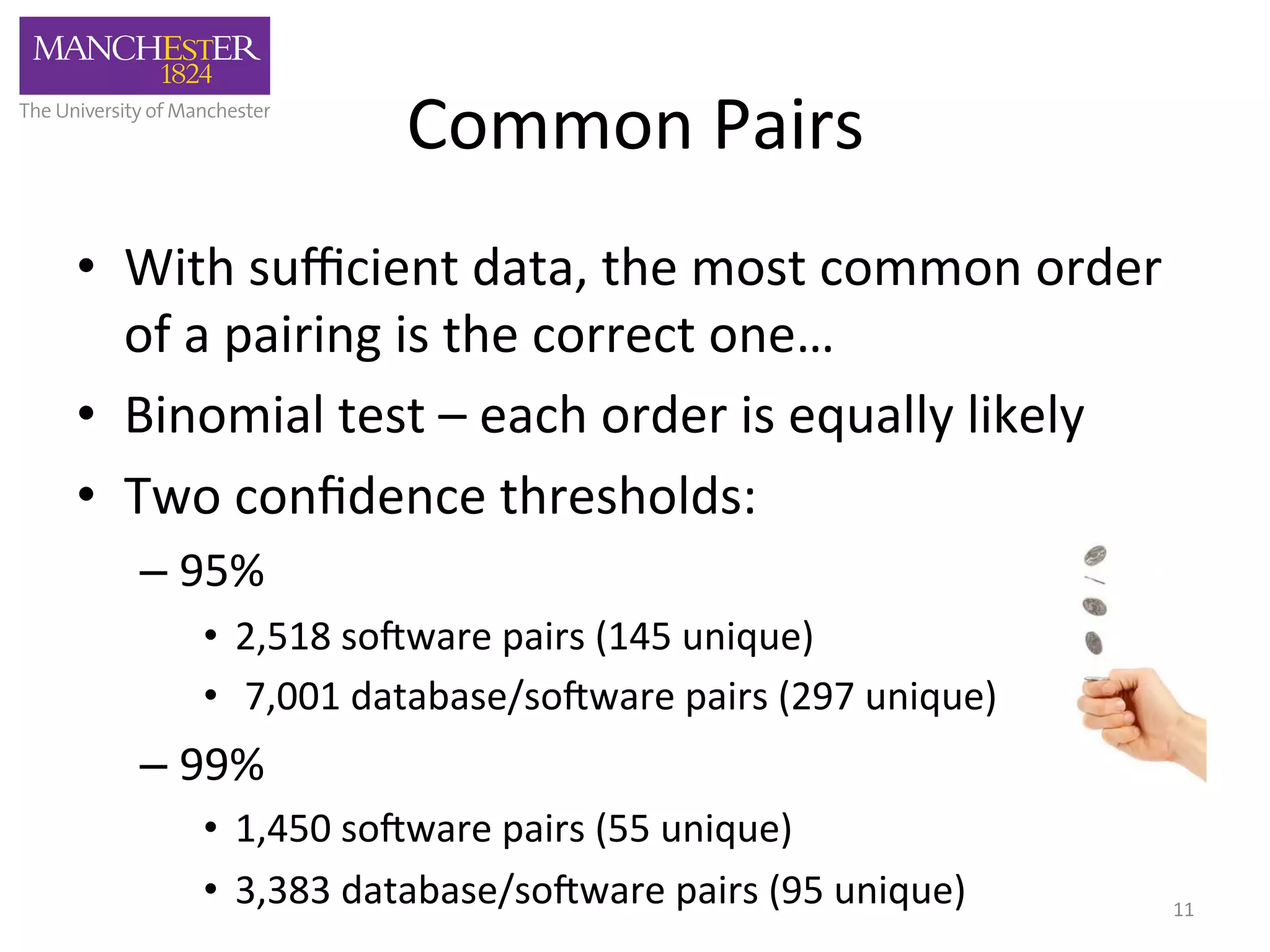
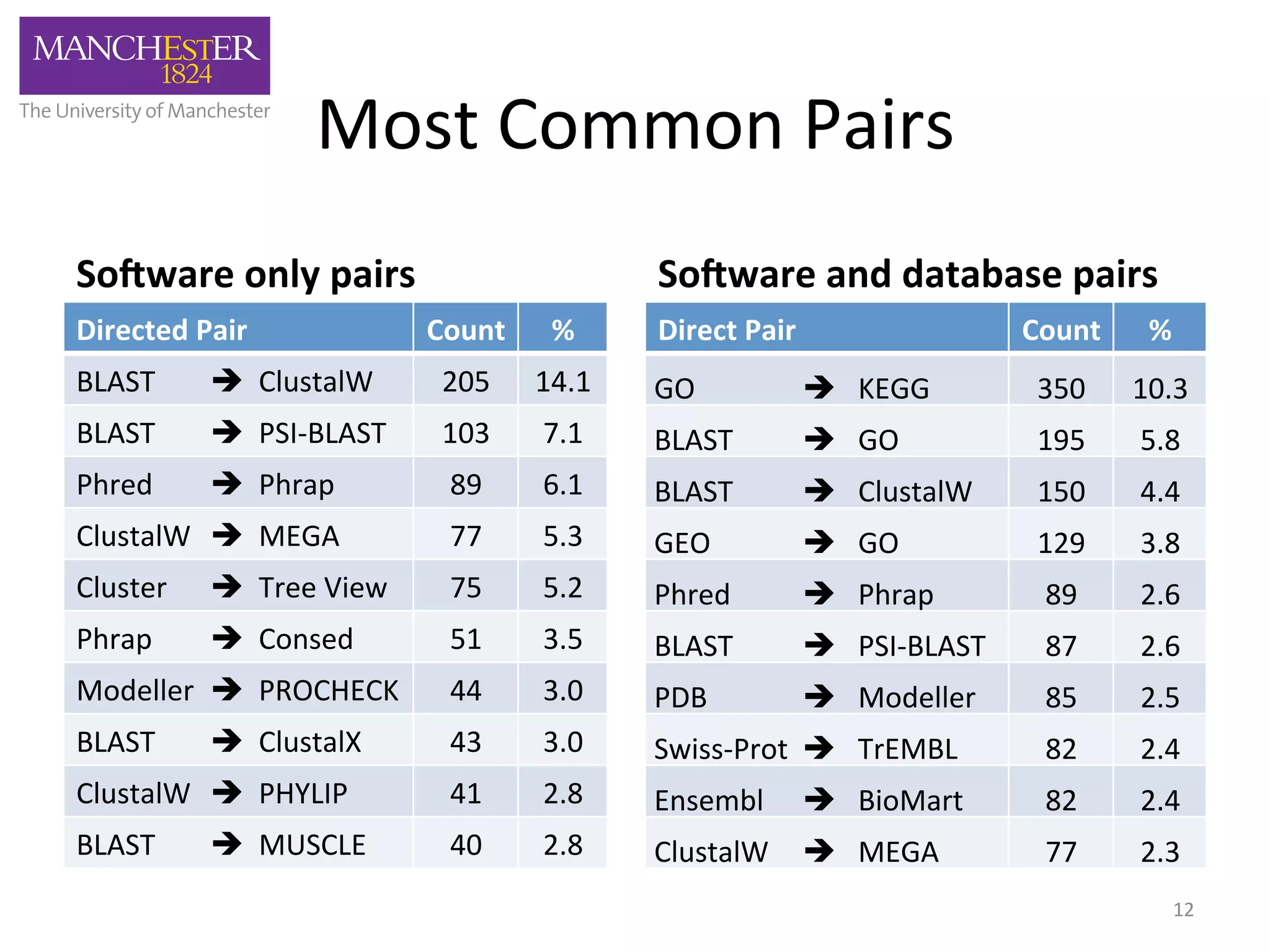
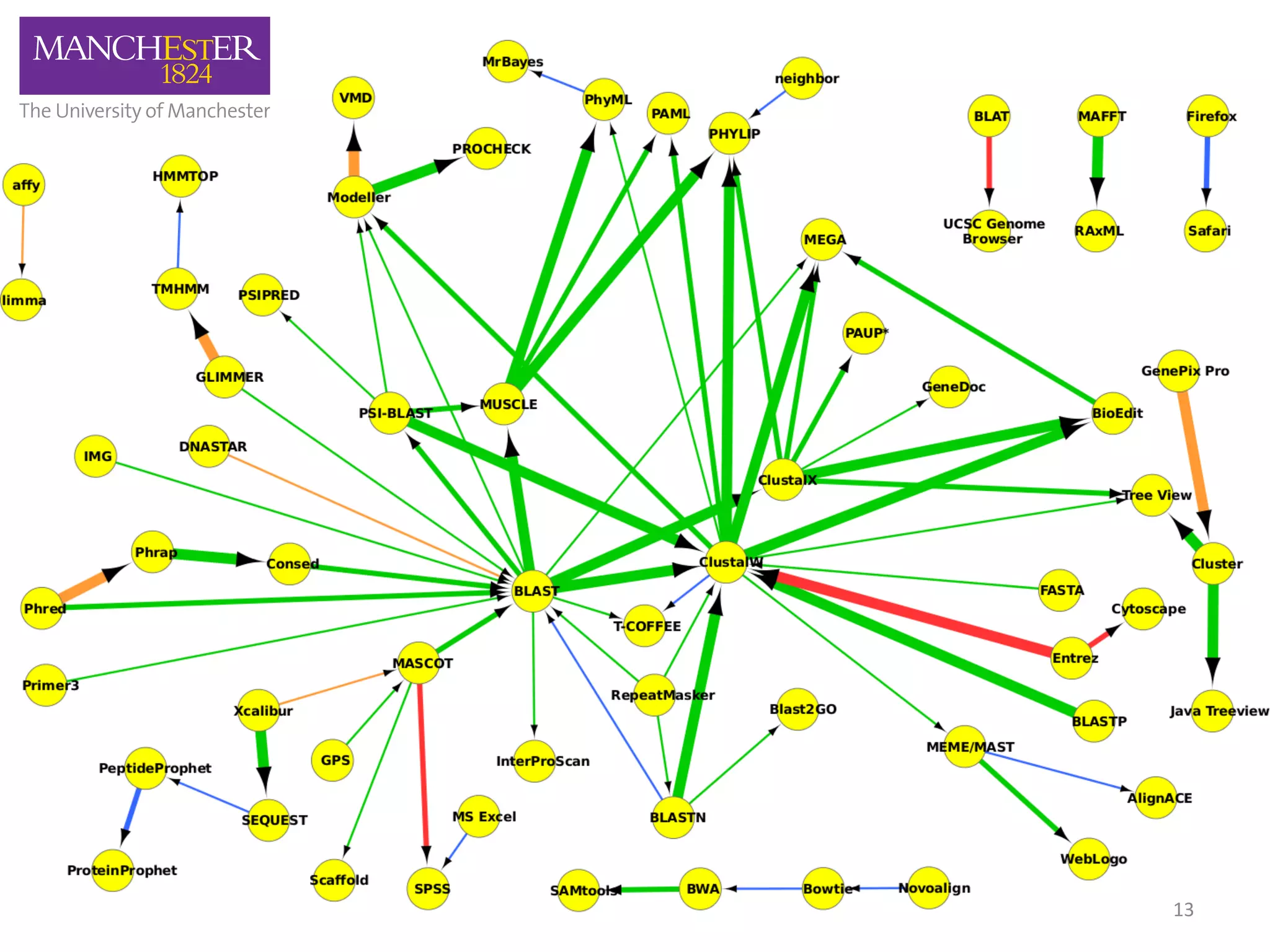
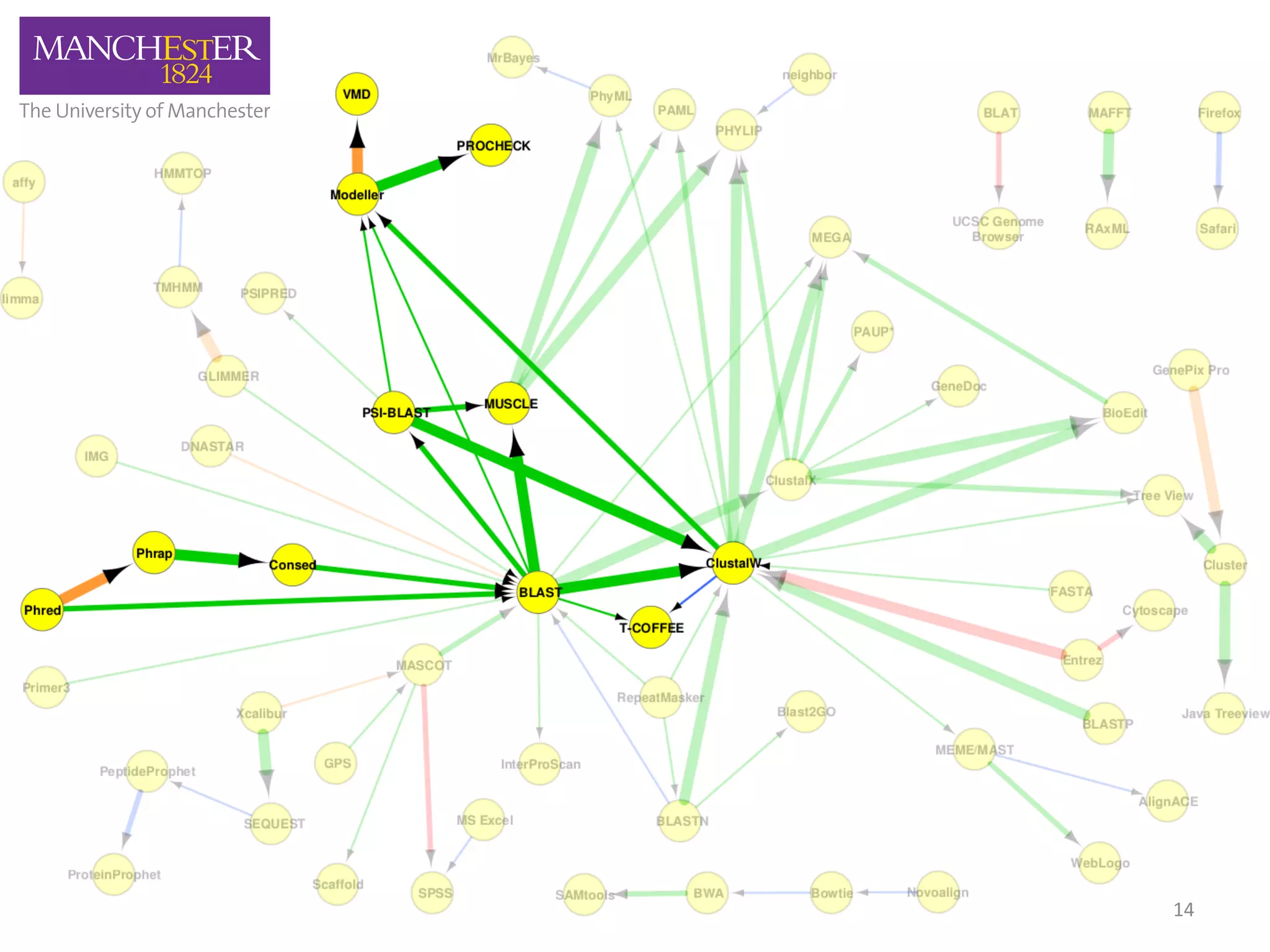
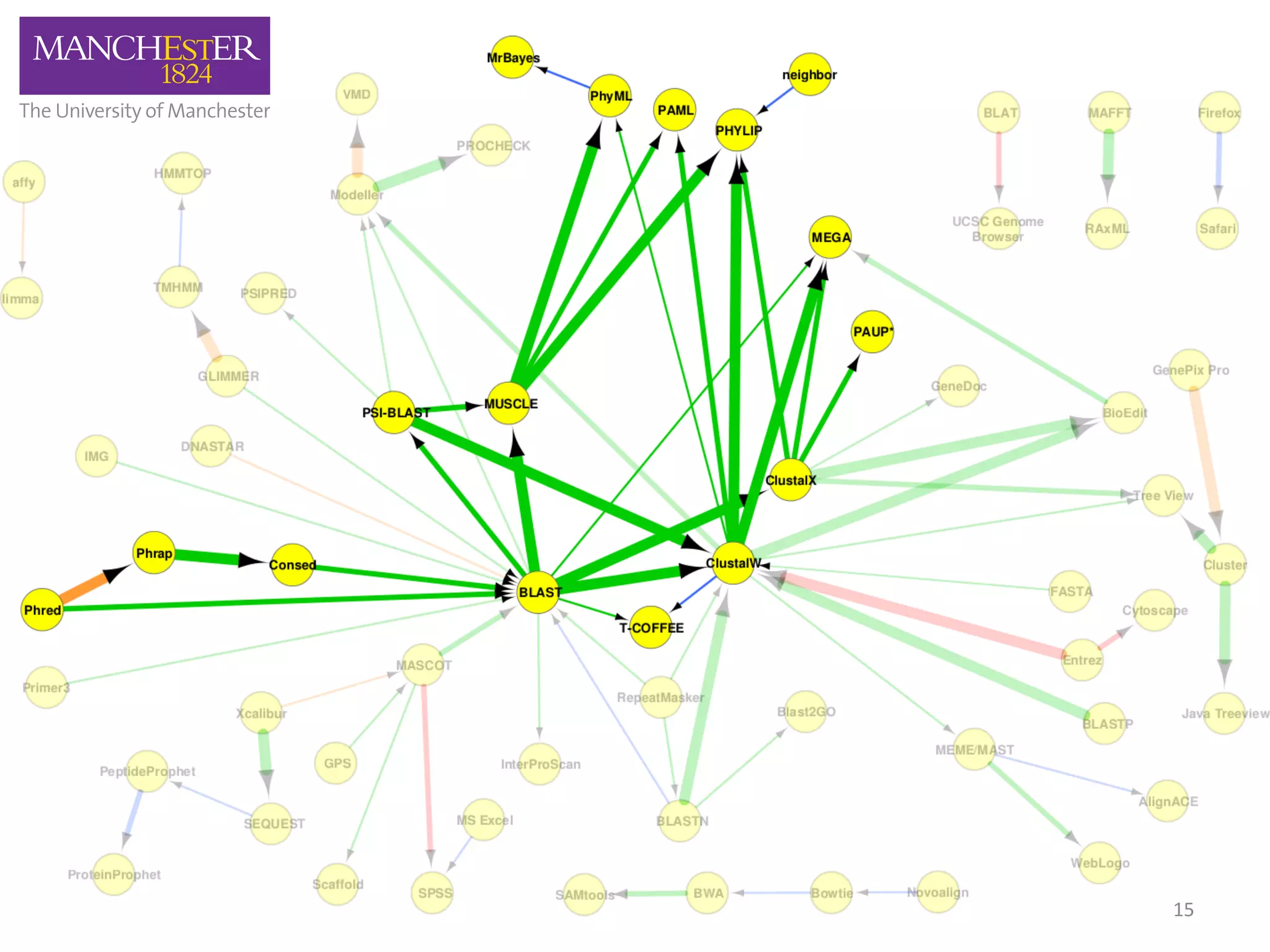
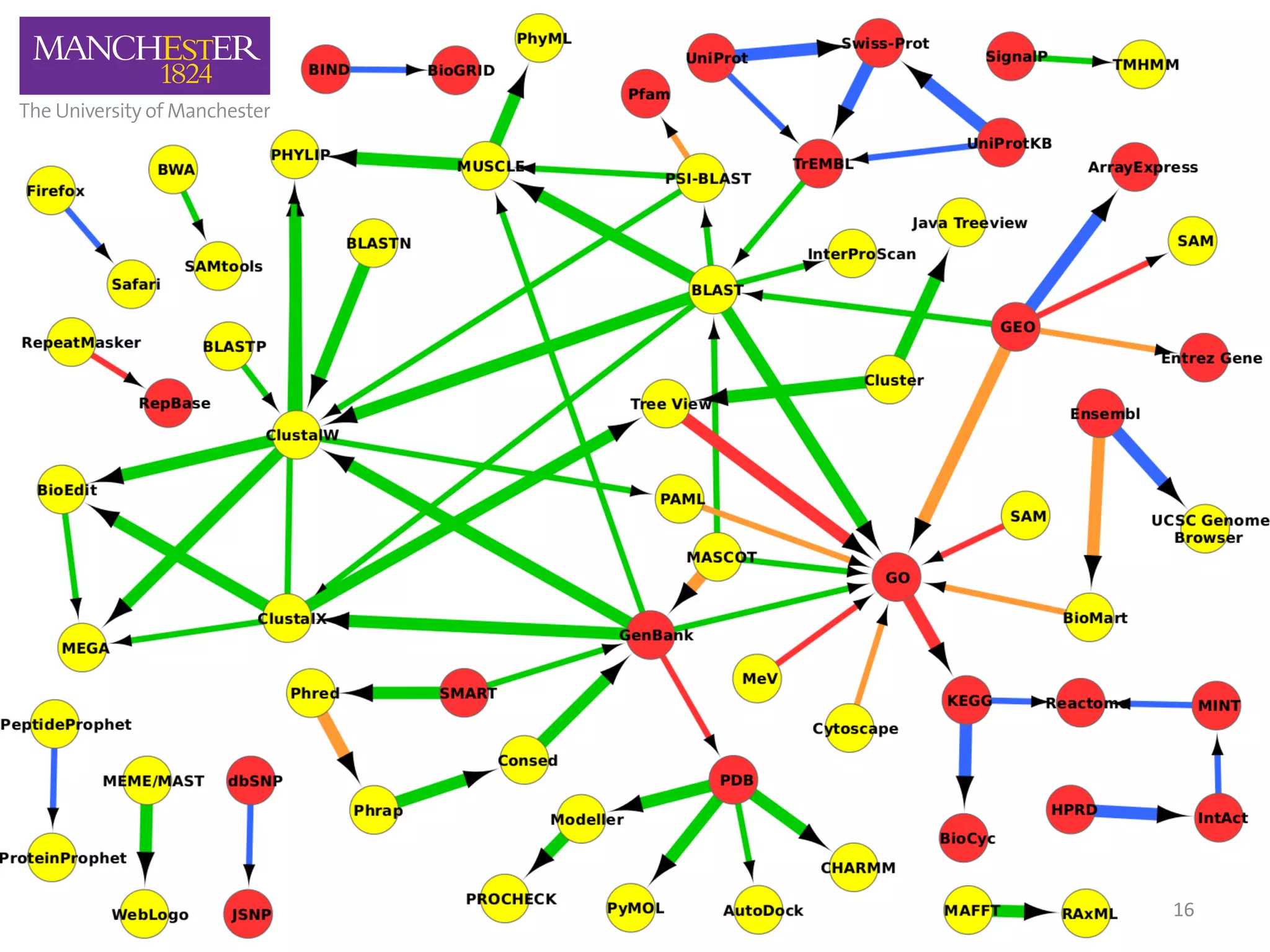
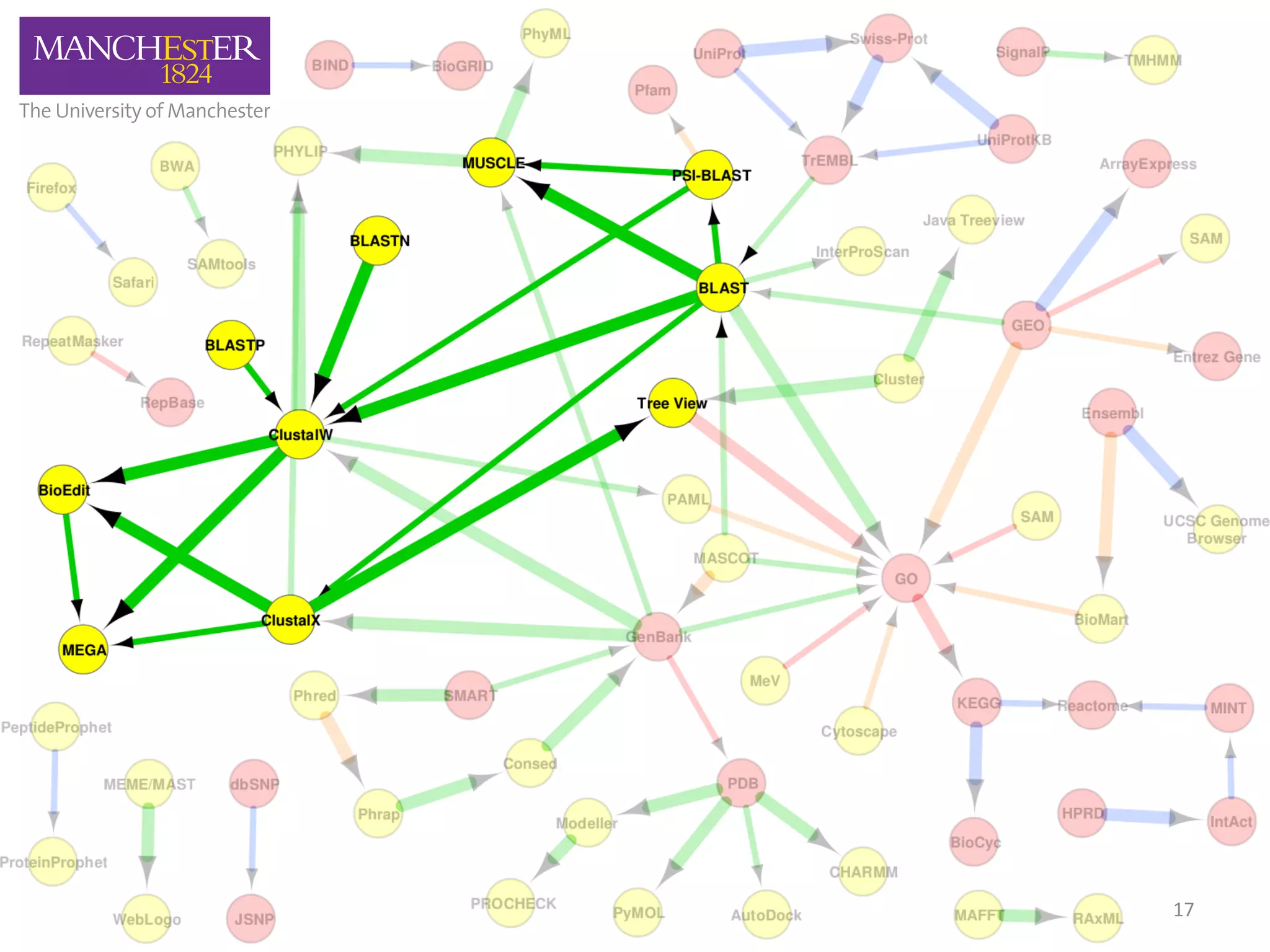
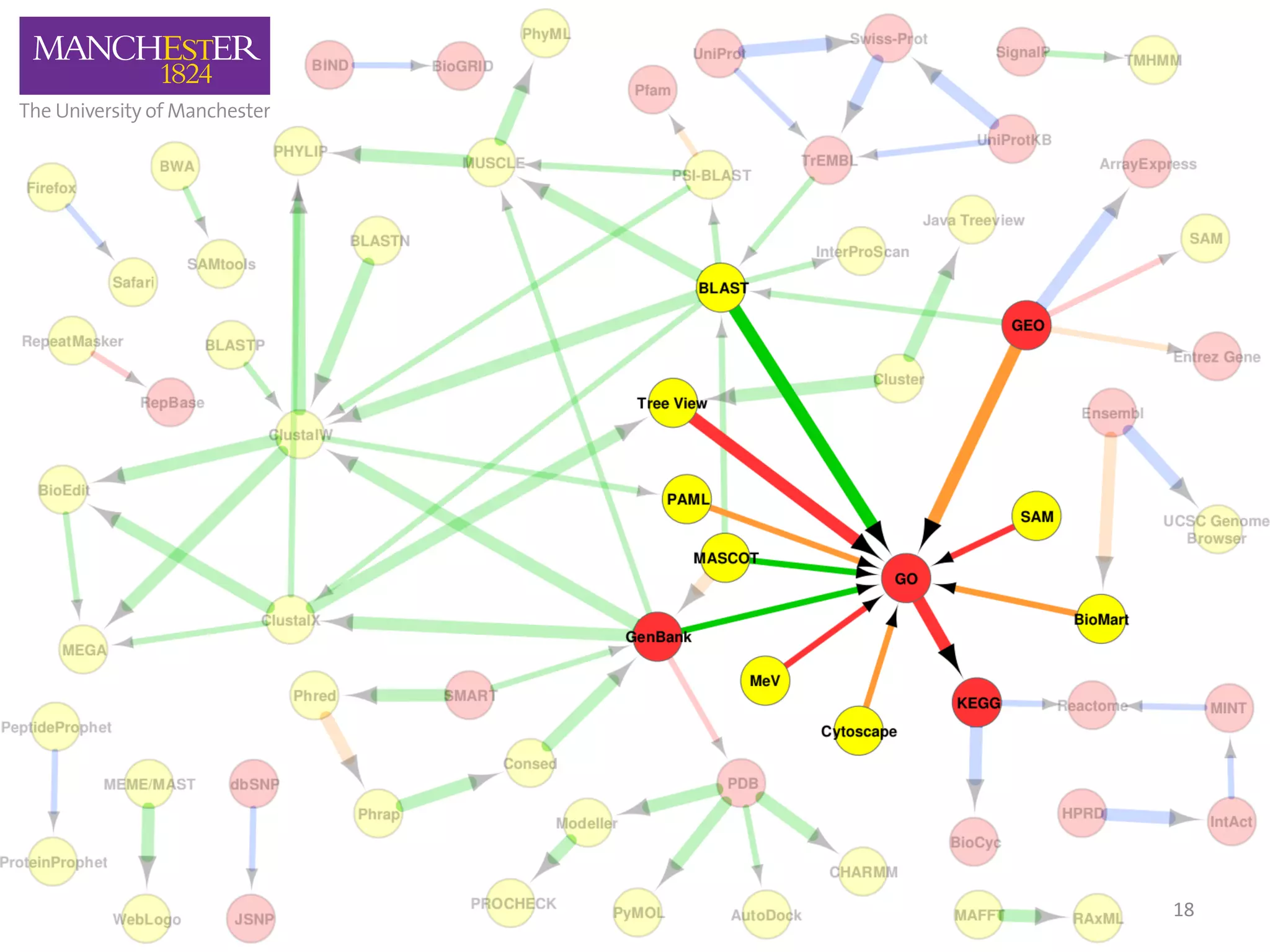
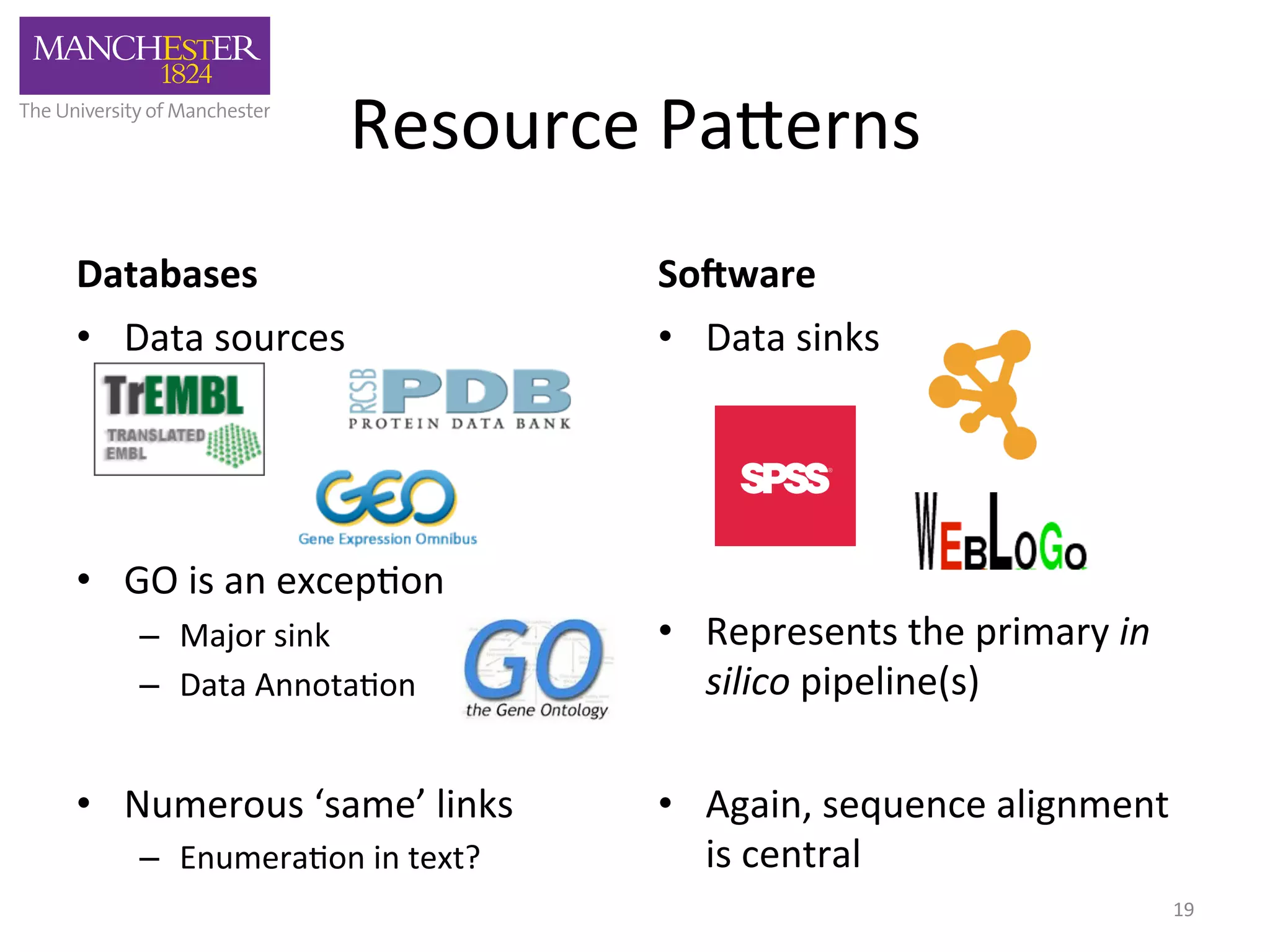
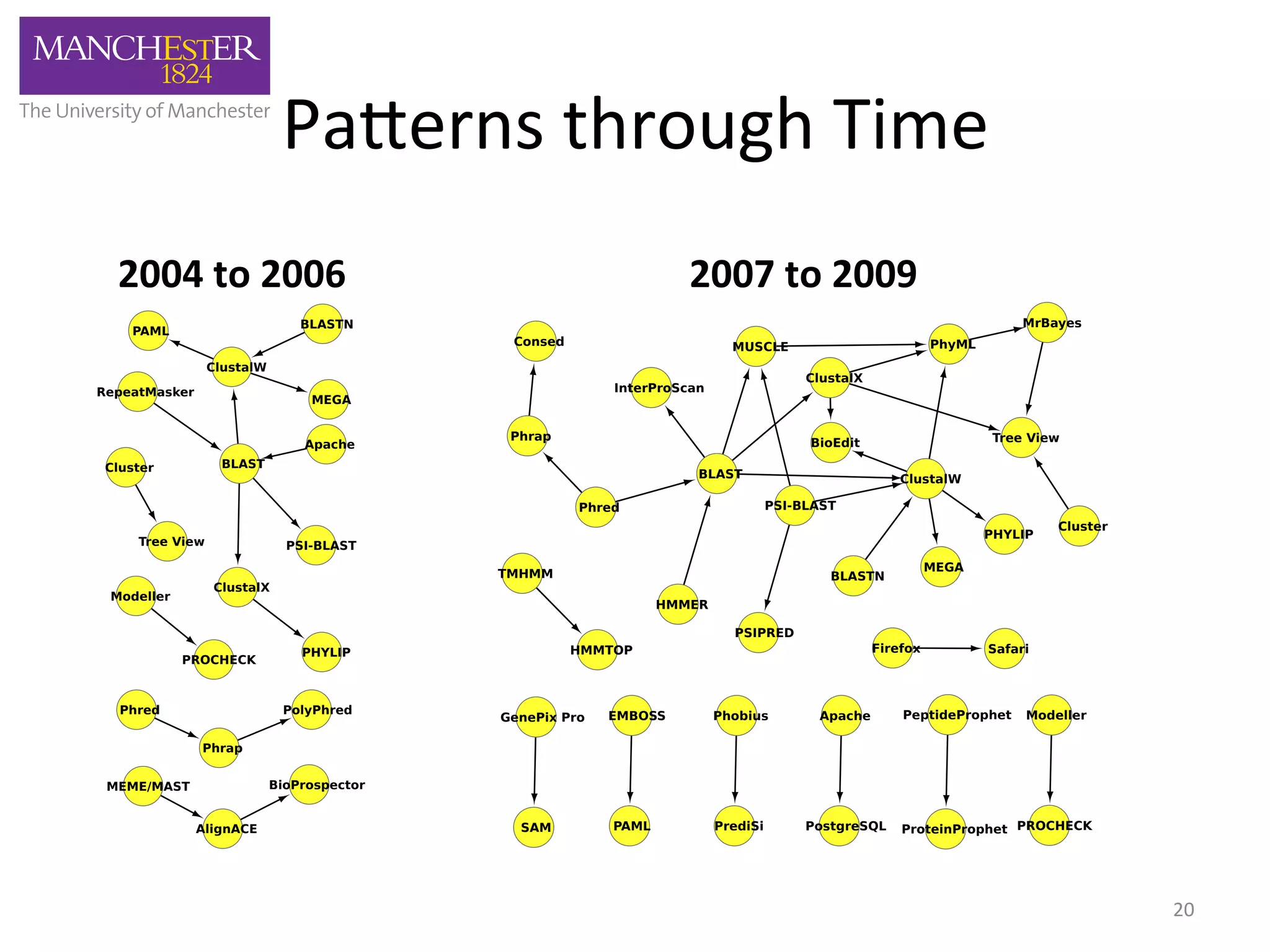
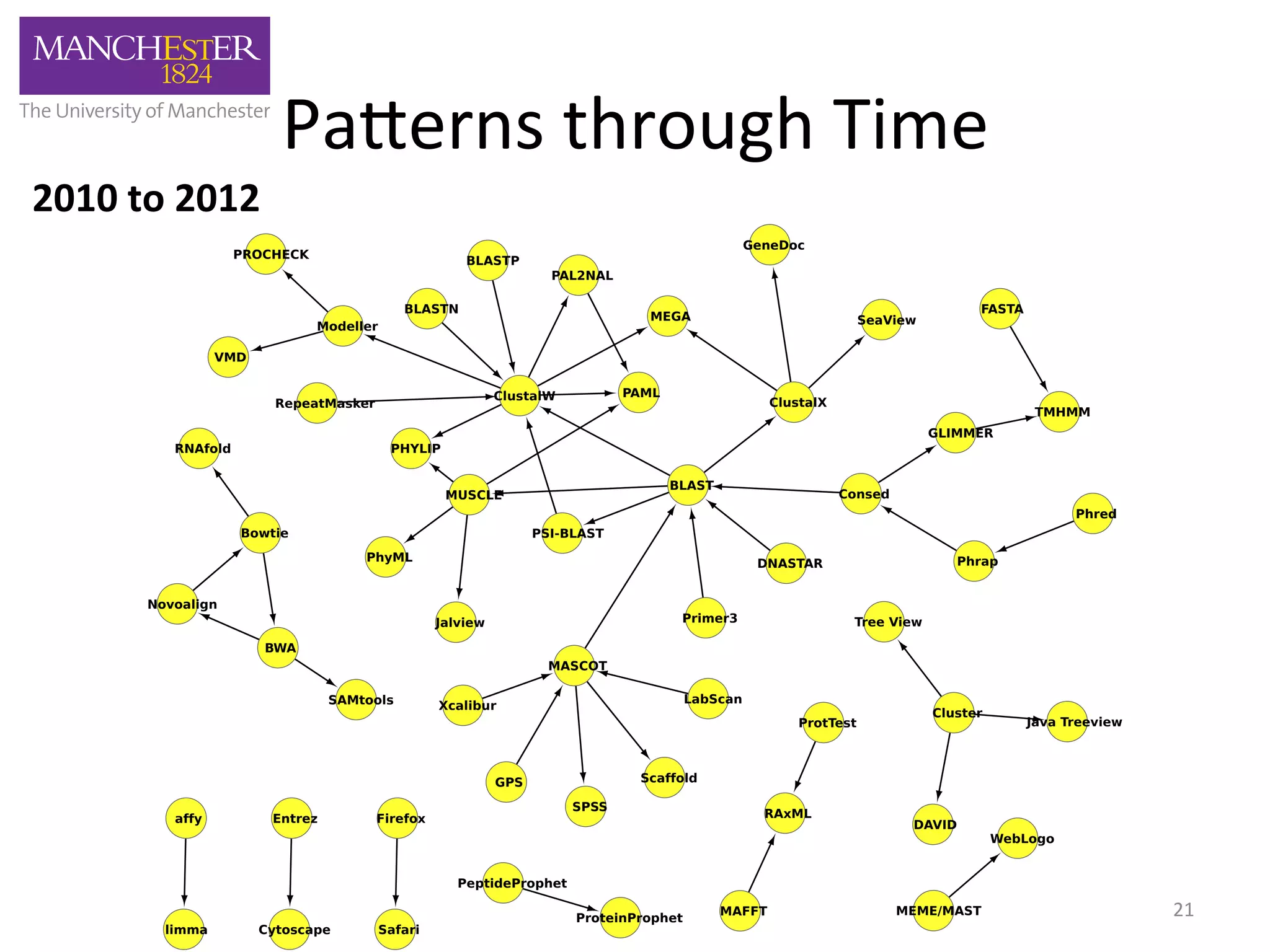
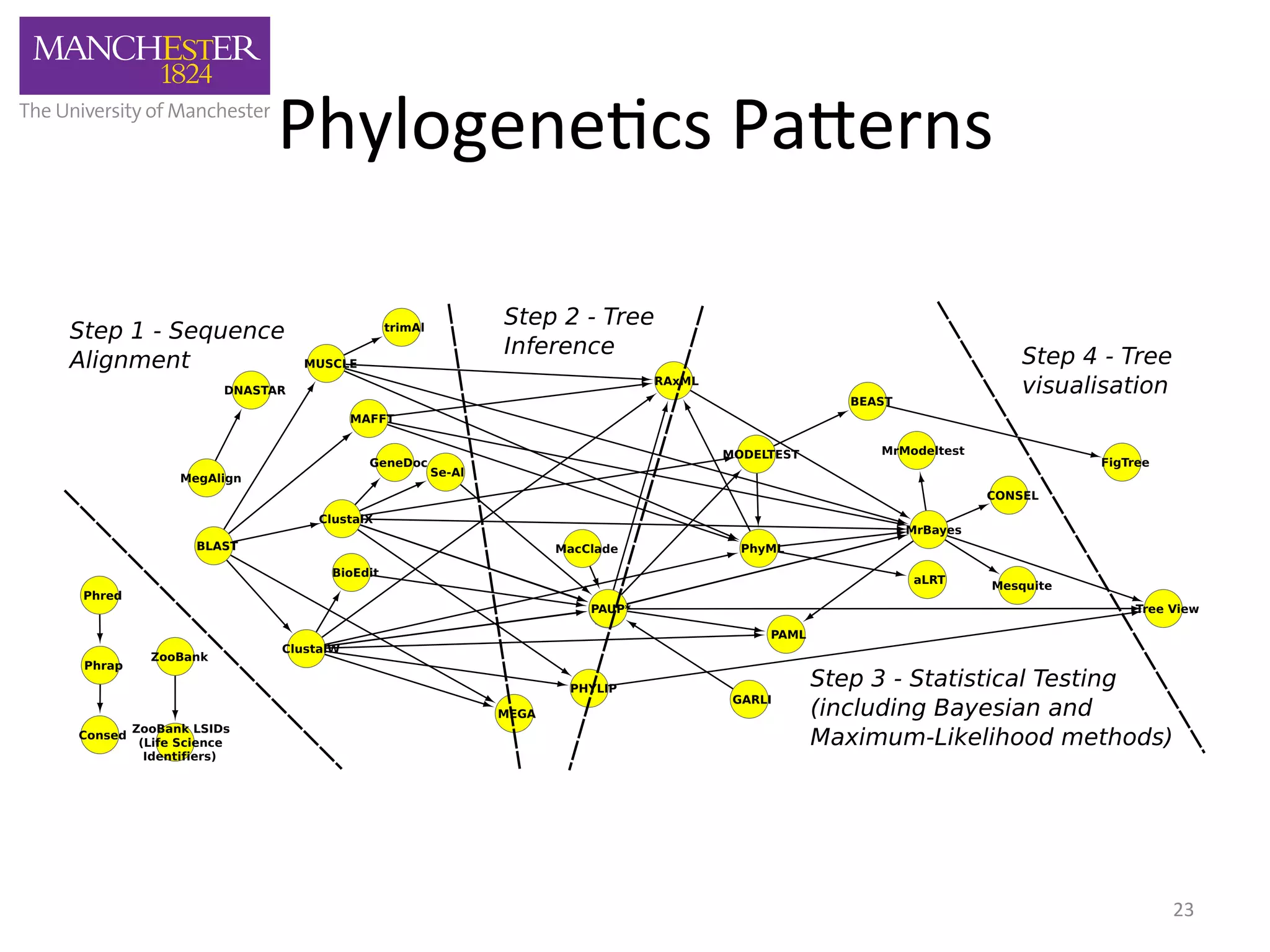
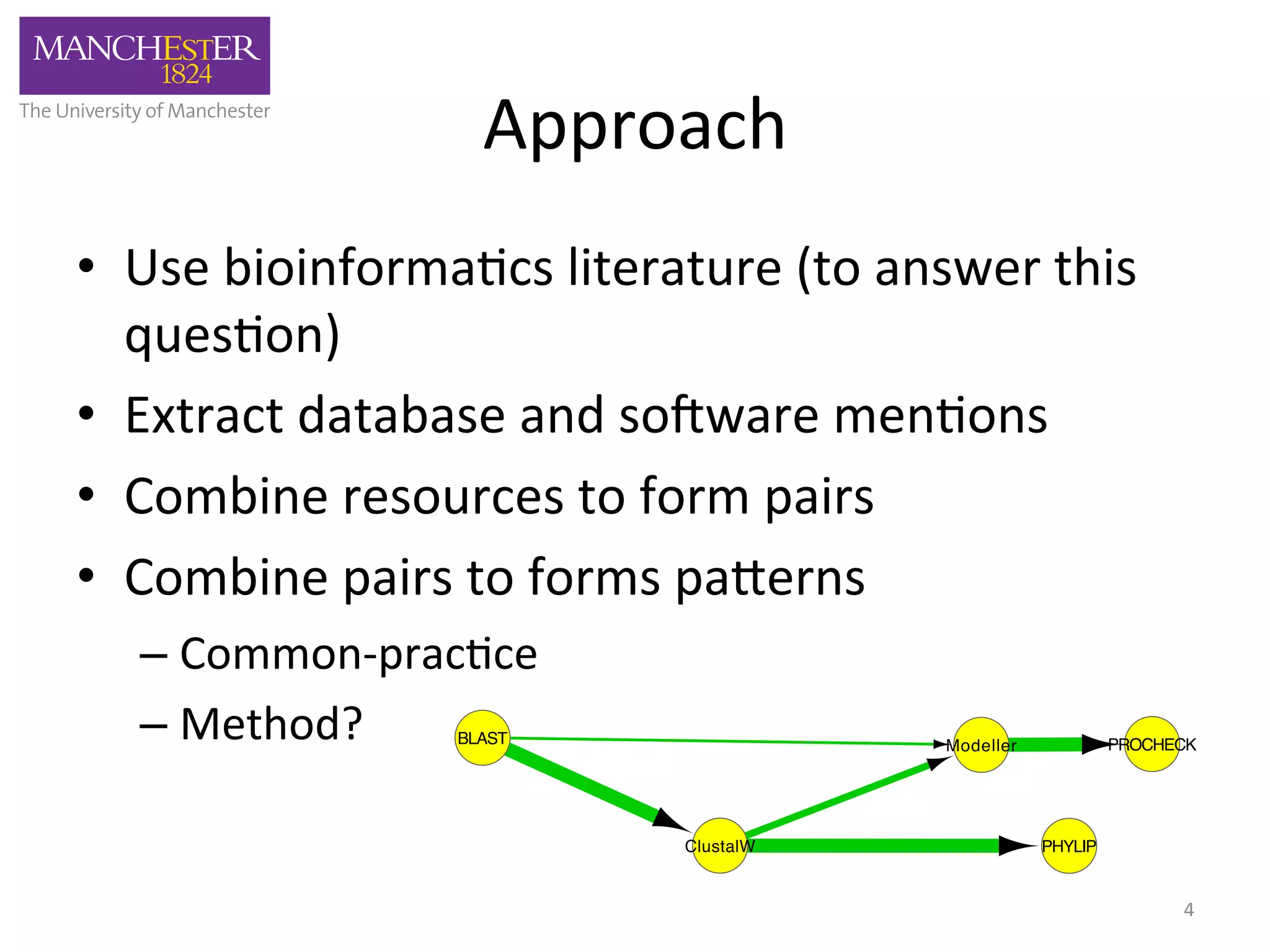
The document discusses the extraction of patterns related to database and software usage in bioinformatics literature, revealing that approximately 200,000 unique resources exist with over 4 million mentions. It utilizes bioinformatics literature to identify and filter relevant resources, categorize them, and build pairs to analyze common practices. The findings indicate significant trends in resource utilization, particularly in sequence alignment and data annotation methods.

![Document
Collec'on
• PubMed
Central
open-‐access
full-‐text
ar'cles
• Bioinforma2cs[MeSH]
• 22,376
ar'cles
• 67
journals
• 3
journals
were
>
50%
of
total
documents
5
(%#!!"
(%!!!"
'%#!!"
'%!!!"
&%#!!"
&%!!!"
$%#!!"
$%!!!"
#!!"
!"
$))*" &!!!" &!!&" &!!(" &!!+" &!!*" &!$!" &!$&" &!$("
!"#$%&'()'*(+"#%,-.'
/%0&'](https://image.slidesharecdn.com/tc4iaiqvrc62lekp7wl6-signature-6cc0d1802337532ed4e6124d874bdb9c61ae8b753d611f431569c37bd608d61c-poli-140912085946-phpapp02/75/ECCB-2014-Extracting-patterns-of-database-and-software-usage-from-the-bioinformatics-literature-5-2048.jpg)