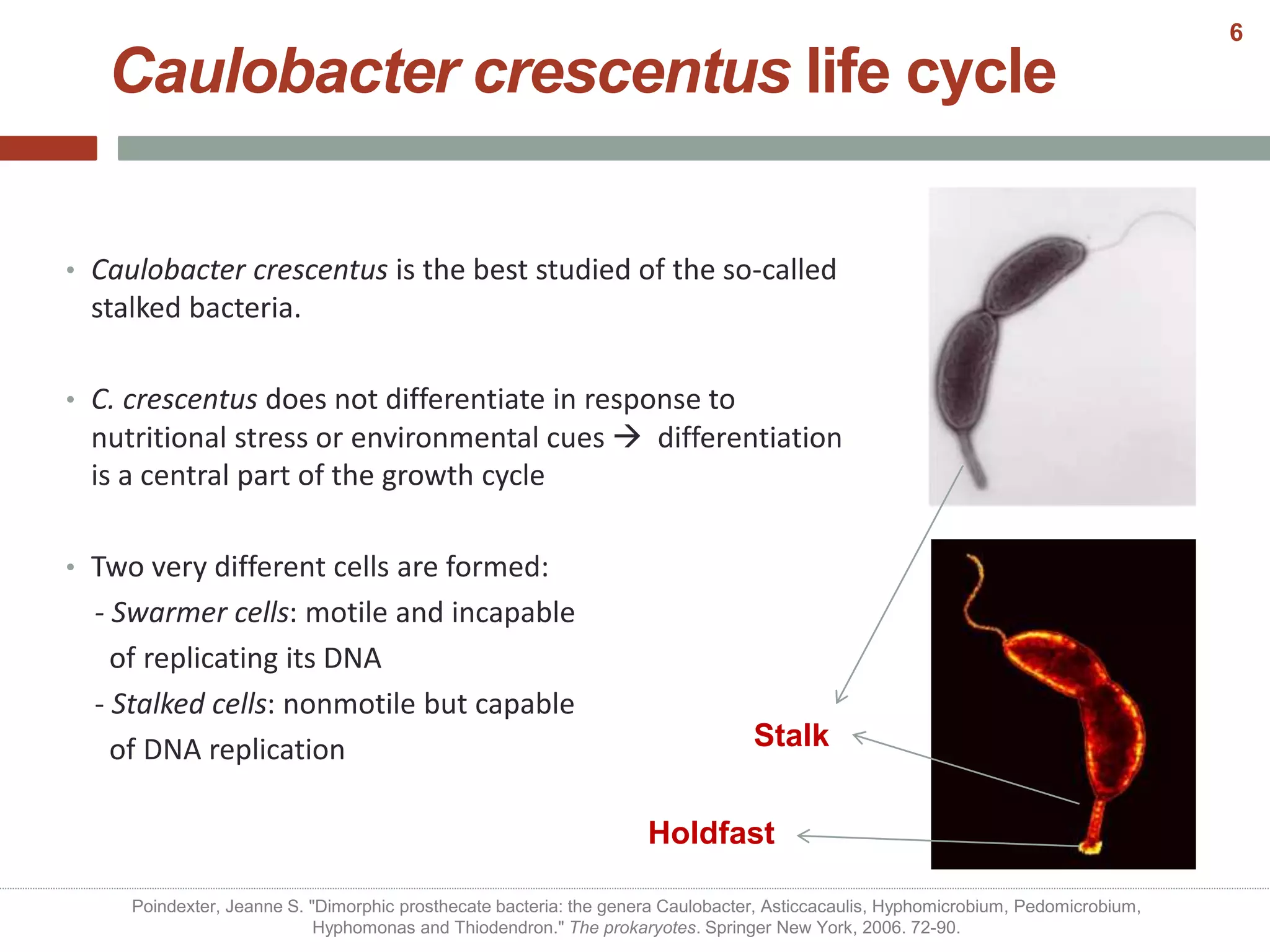
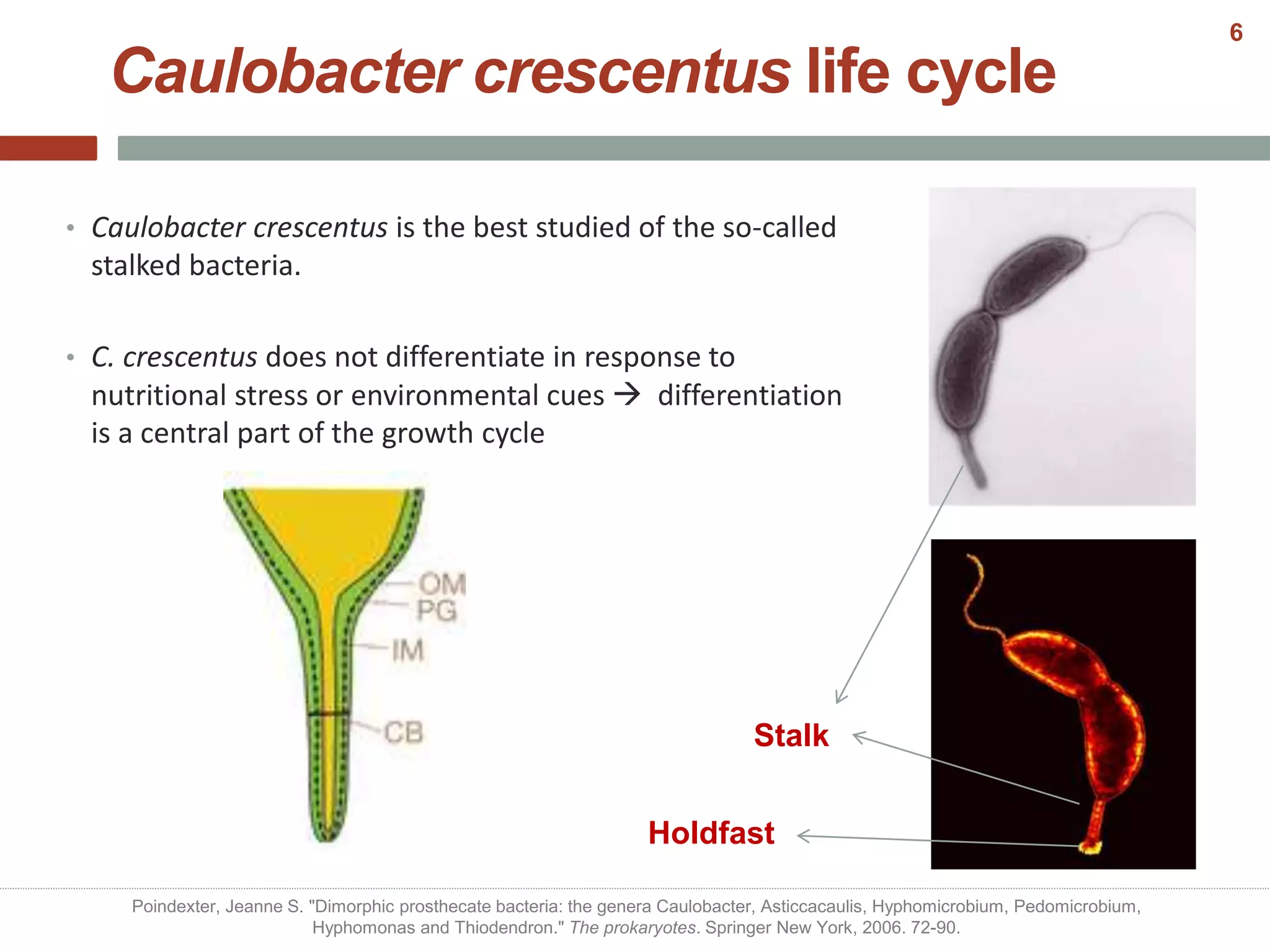
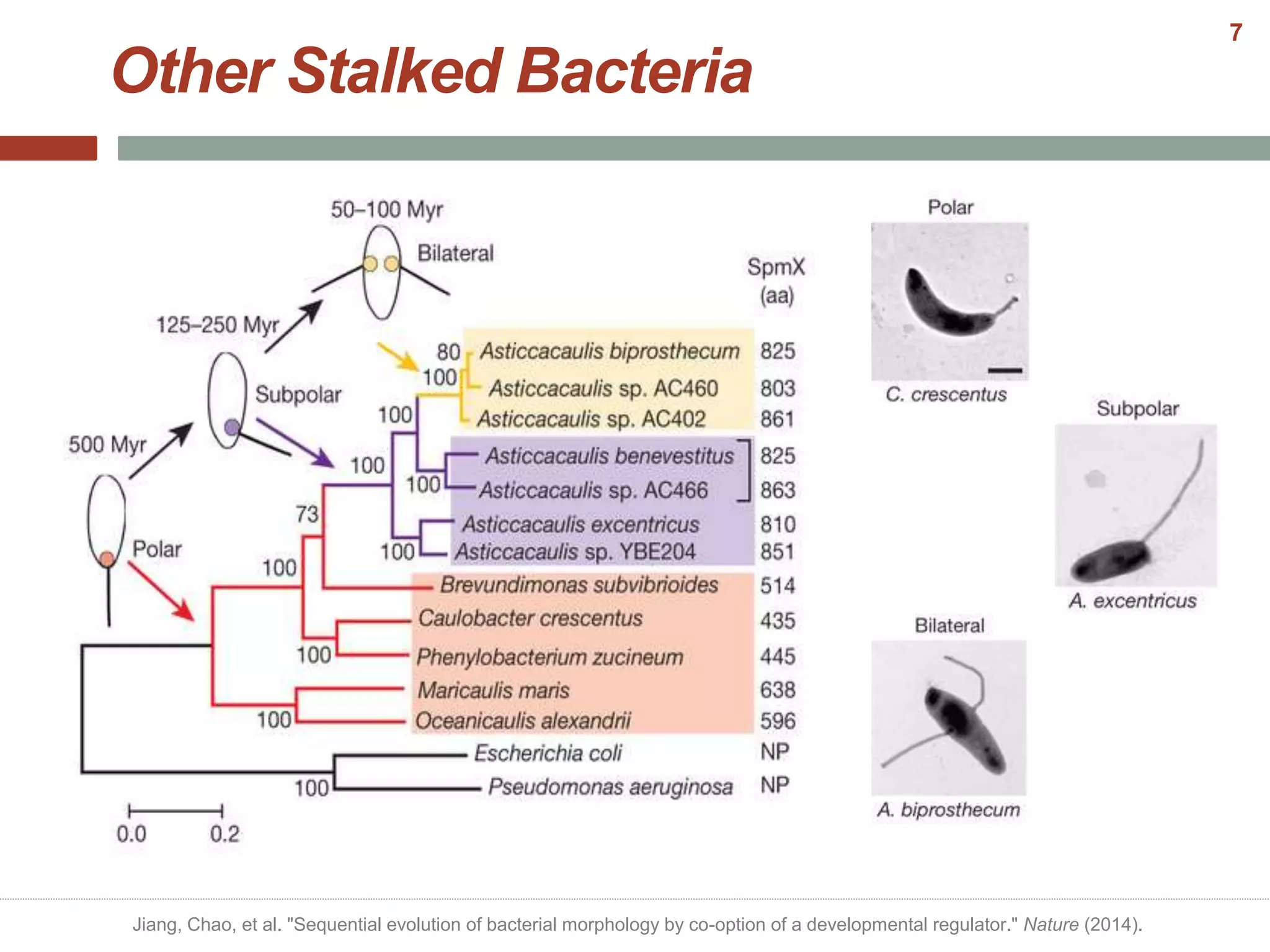
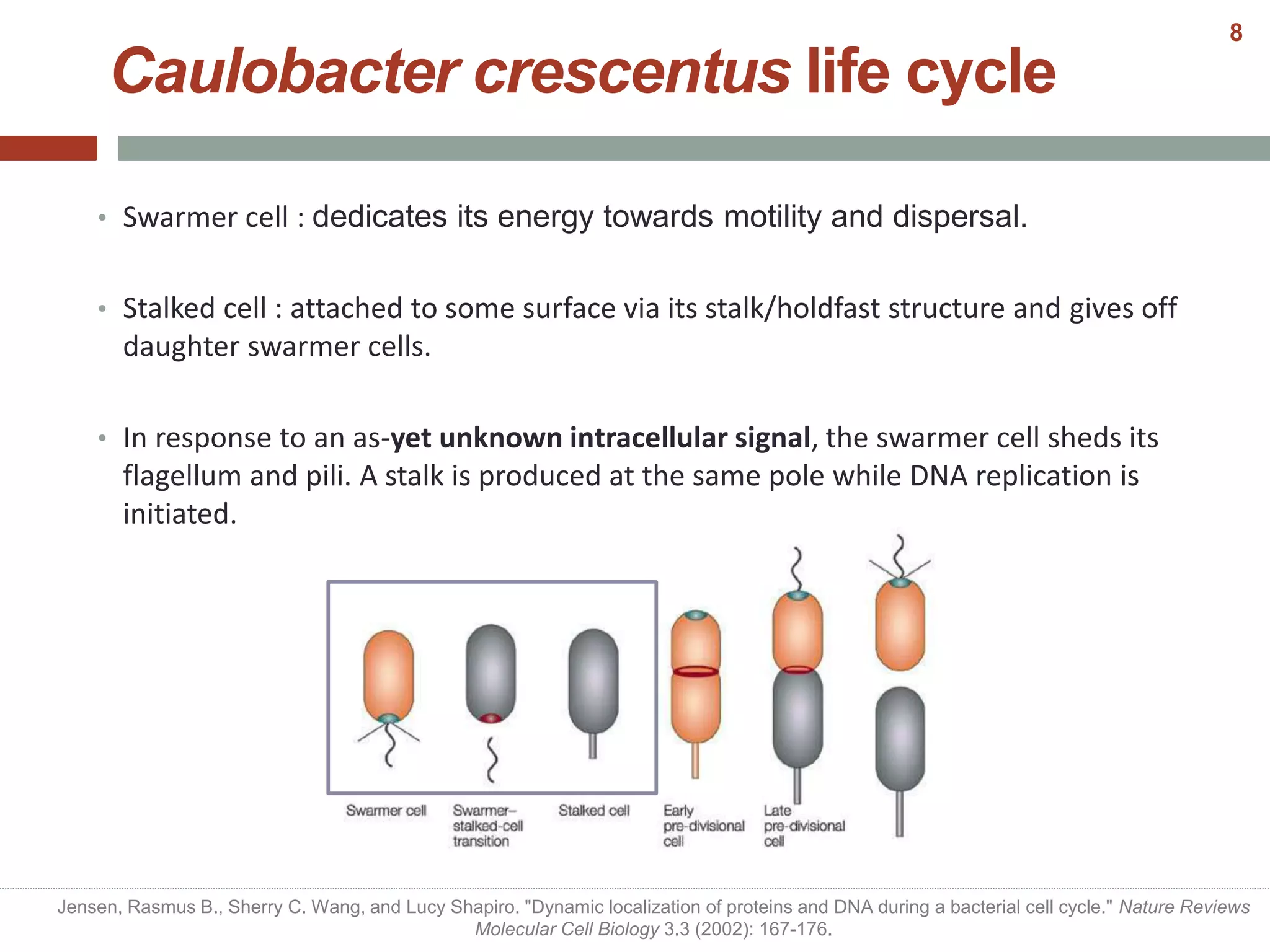
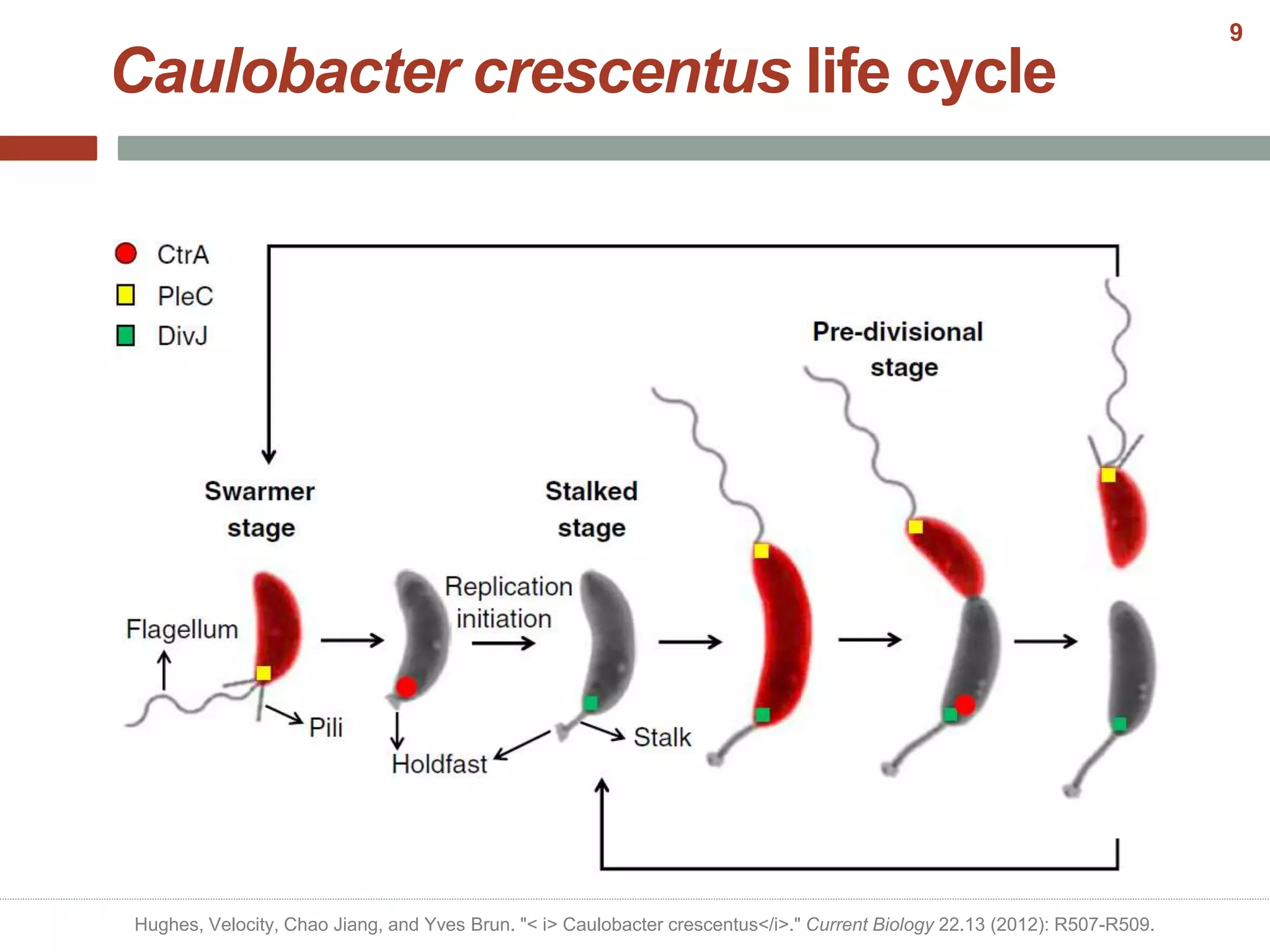
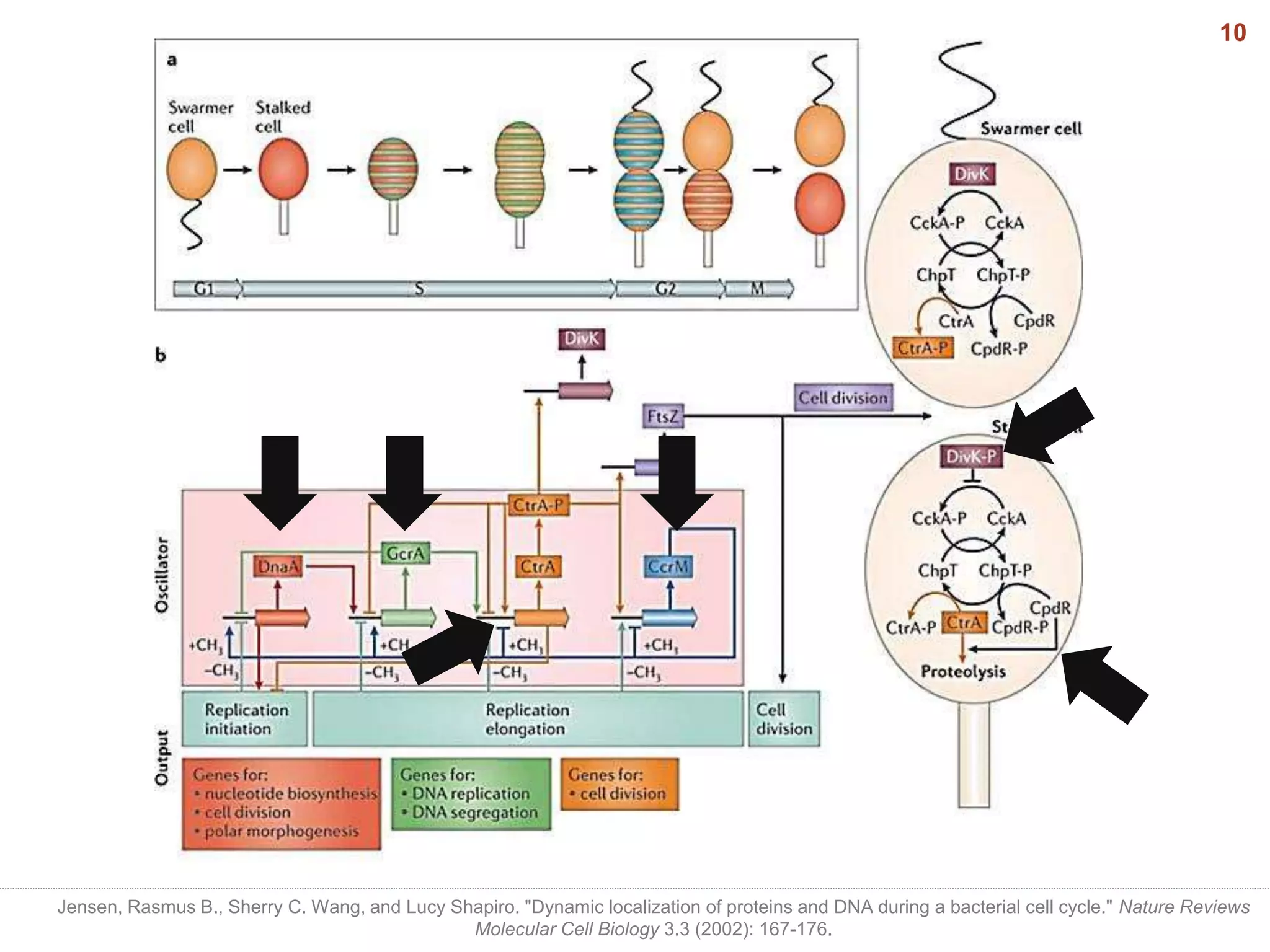
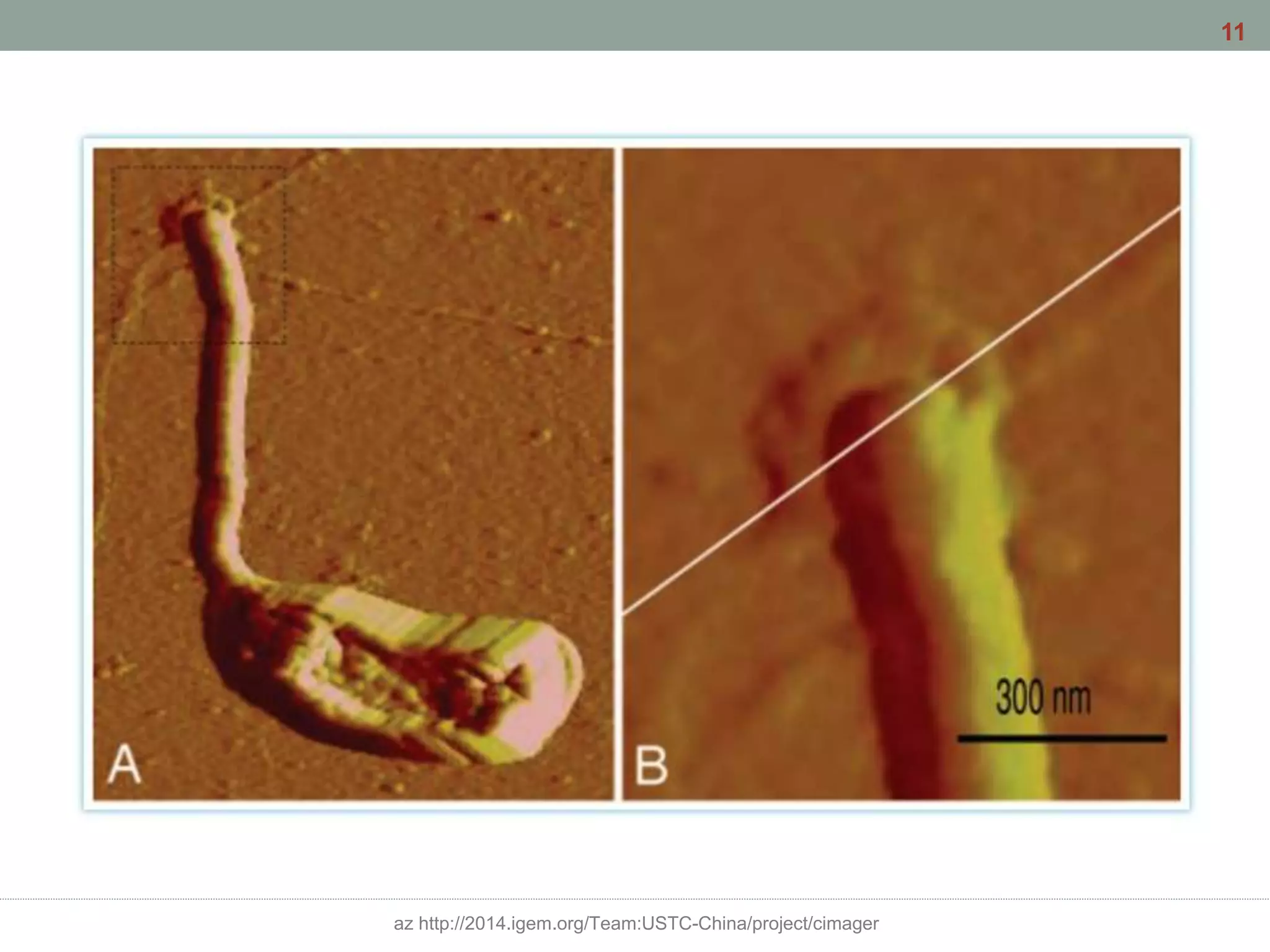
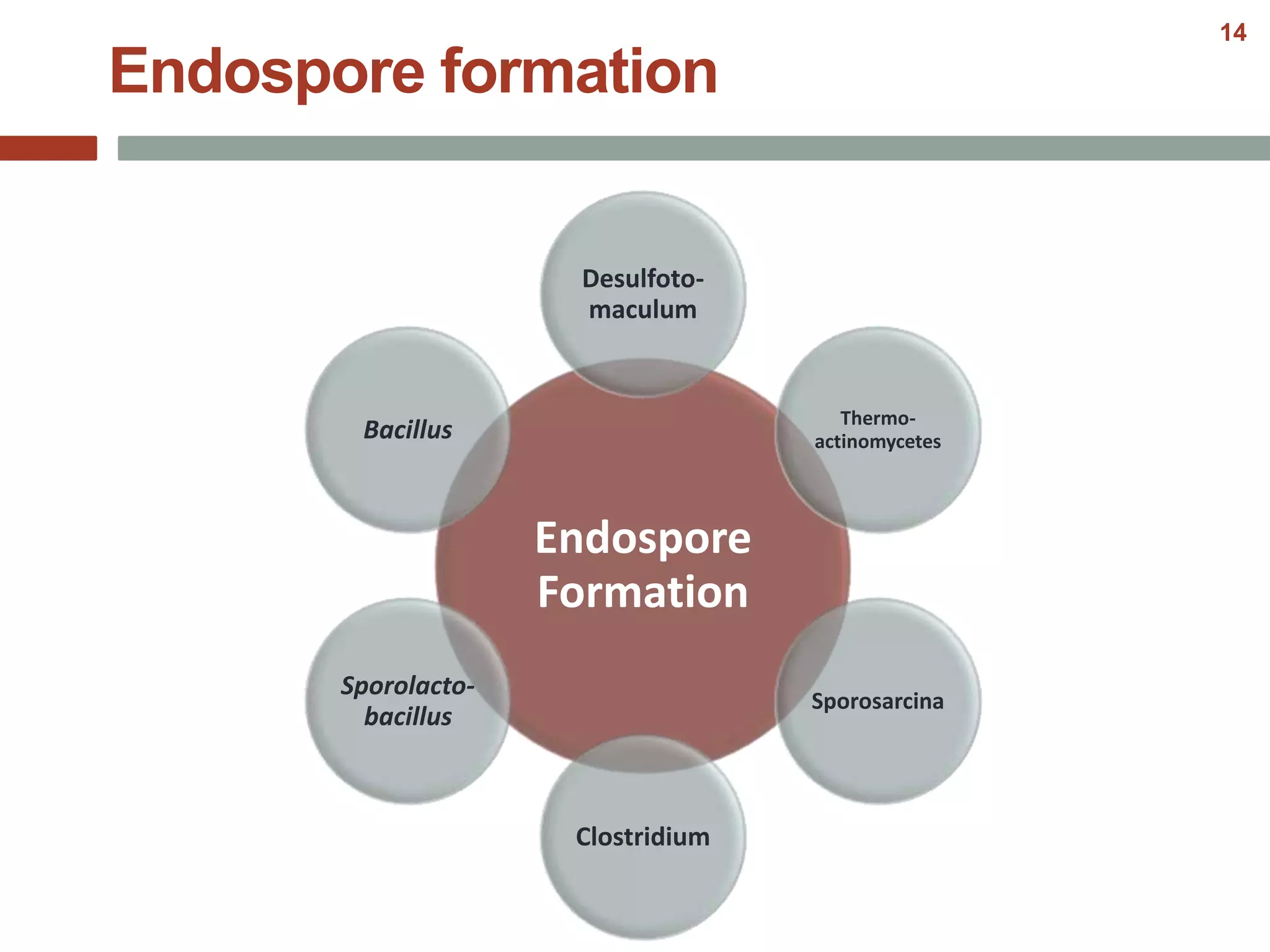
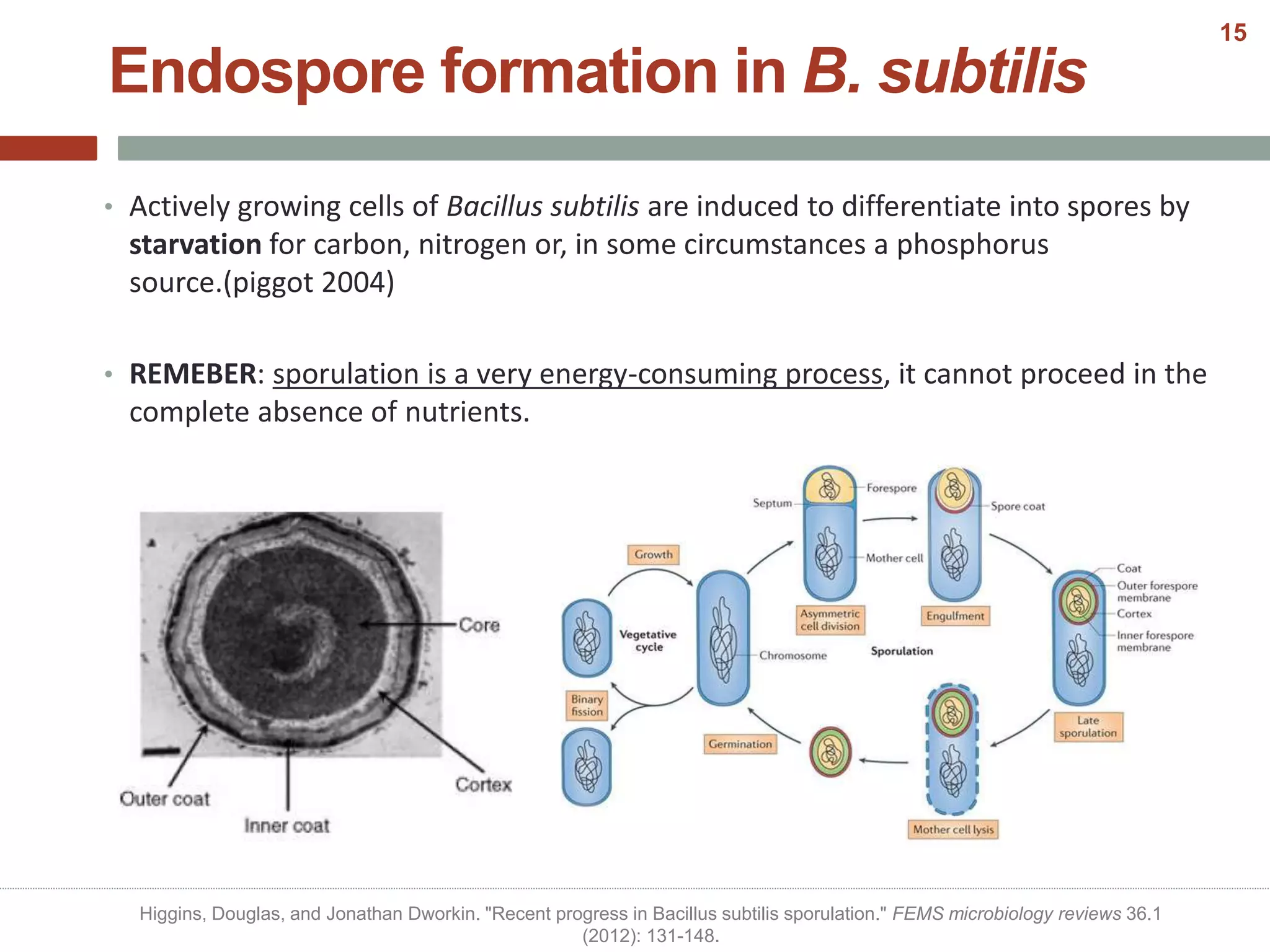
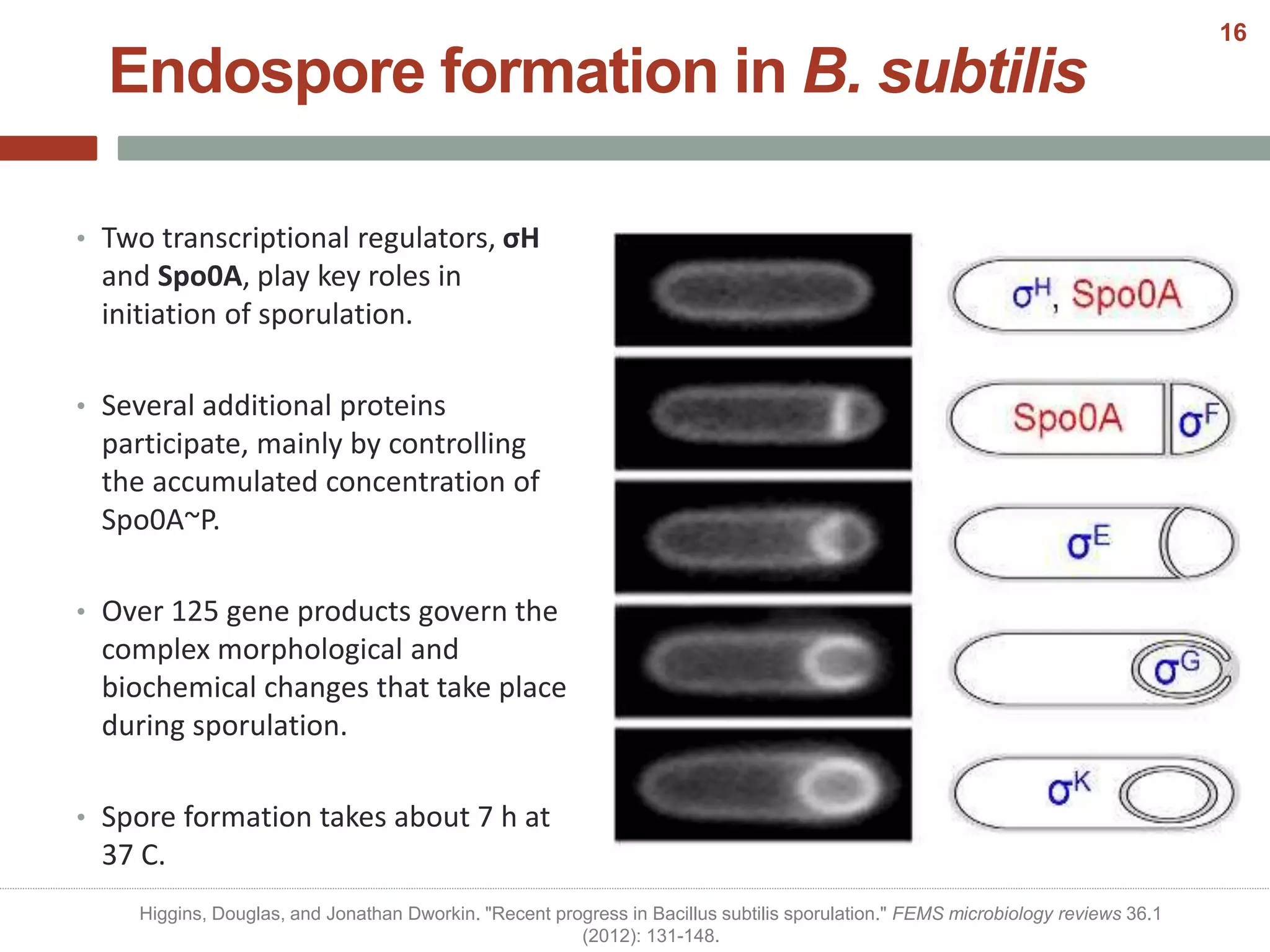
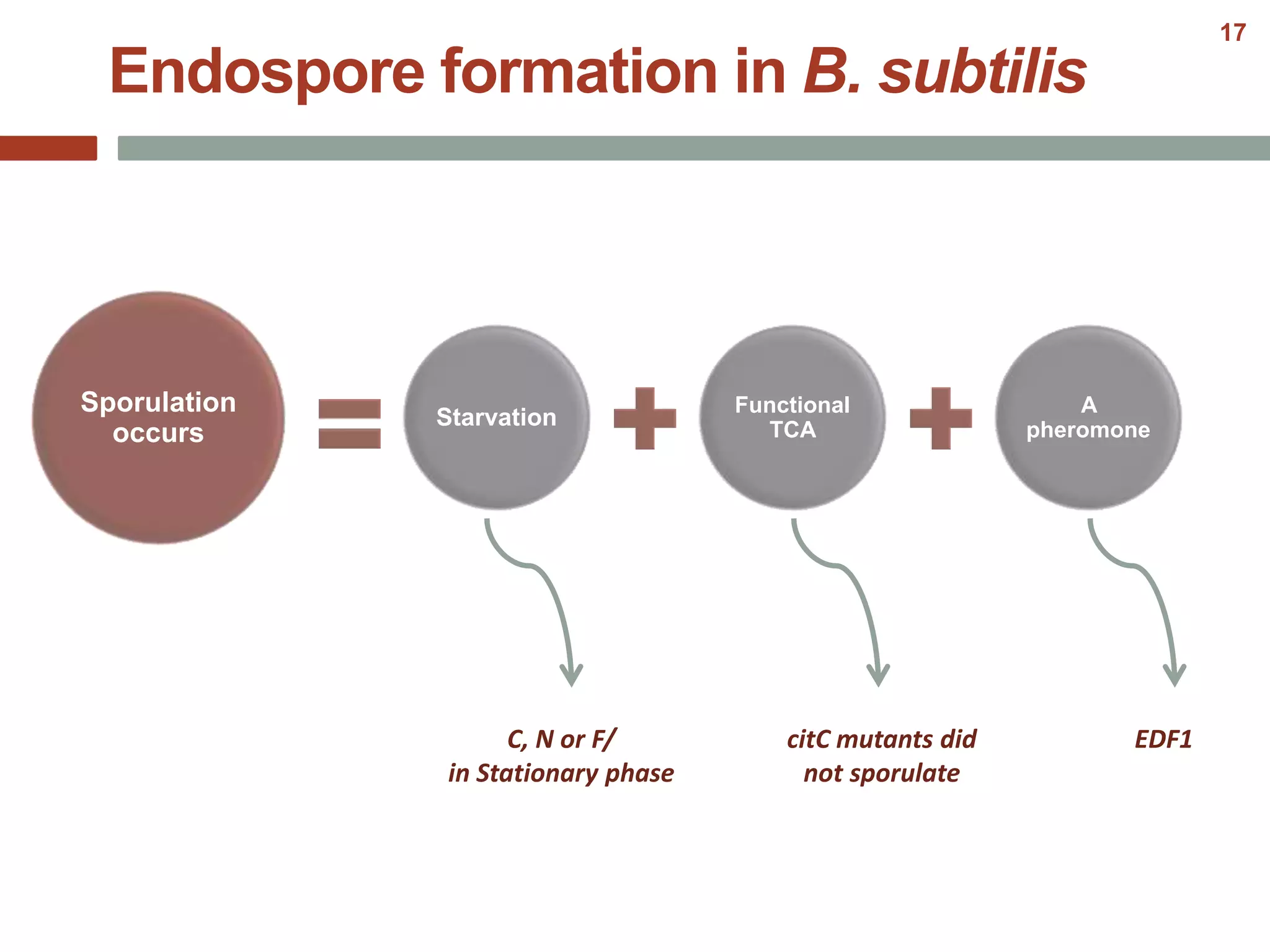
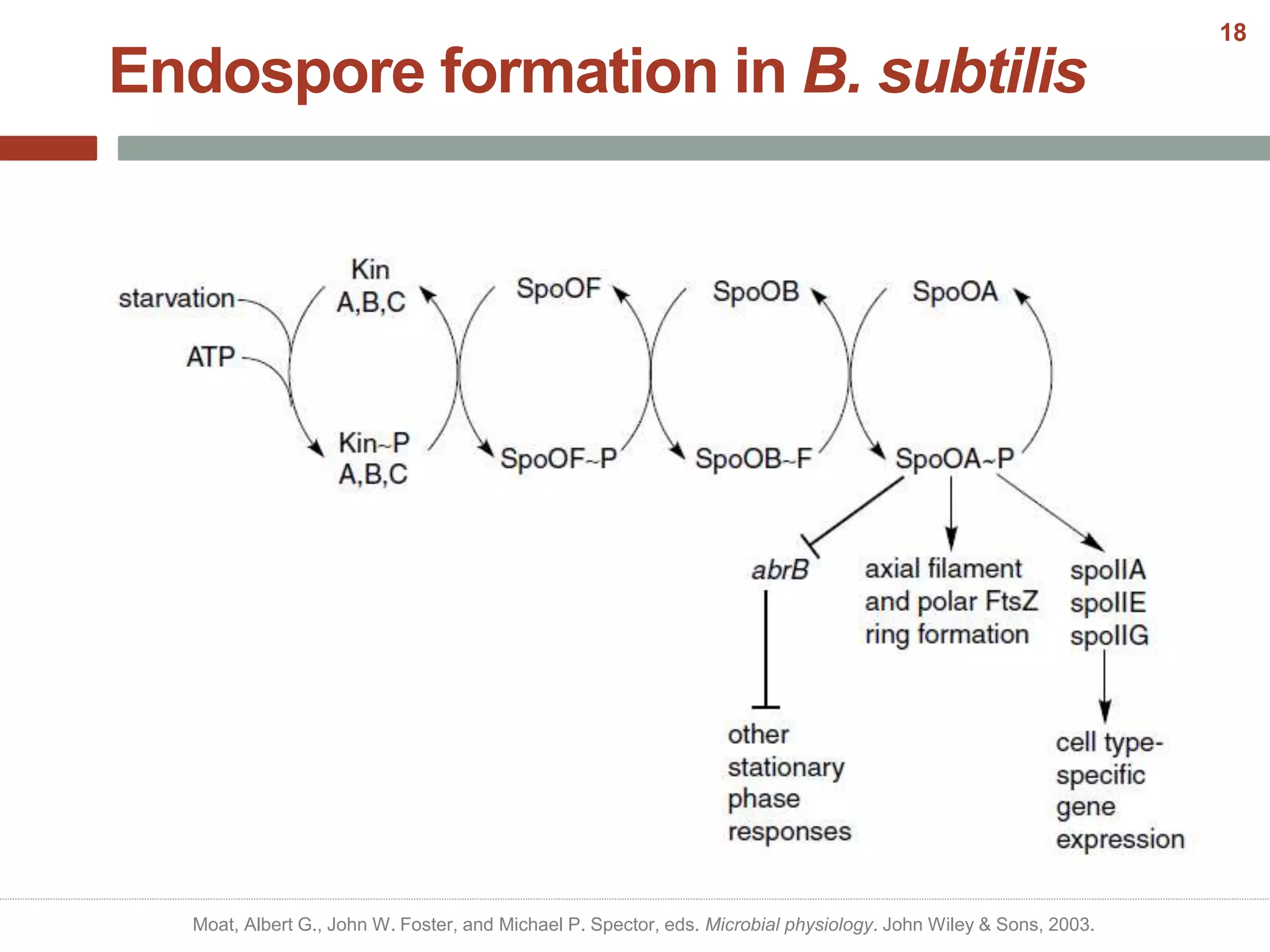
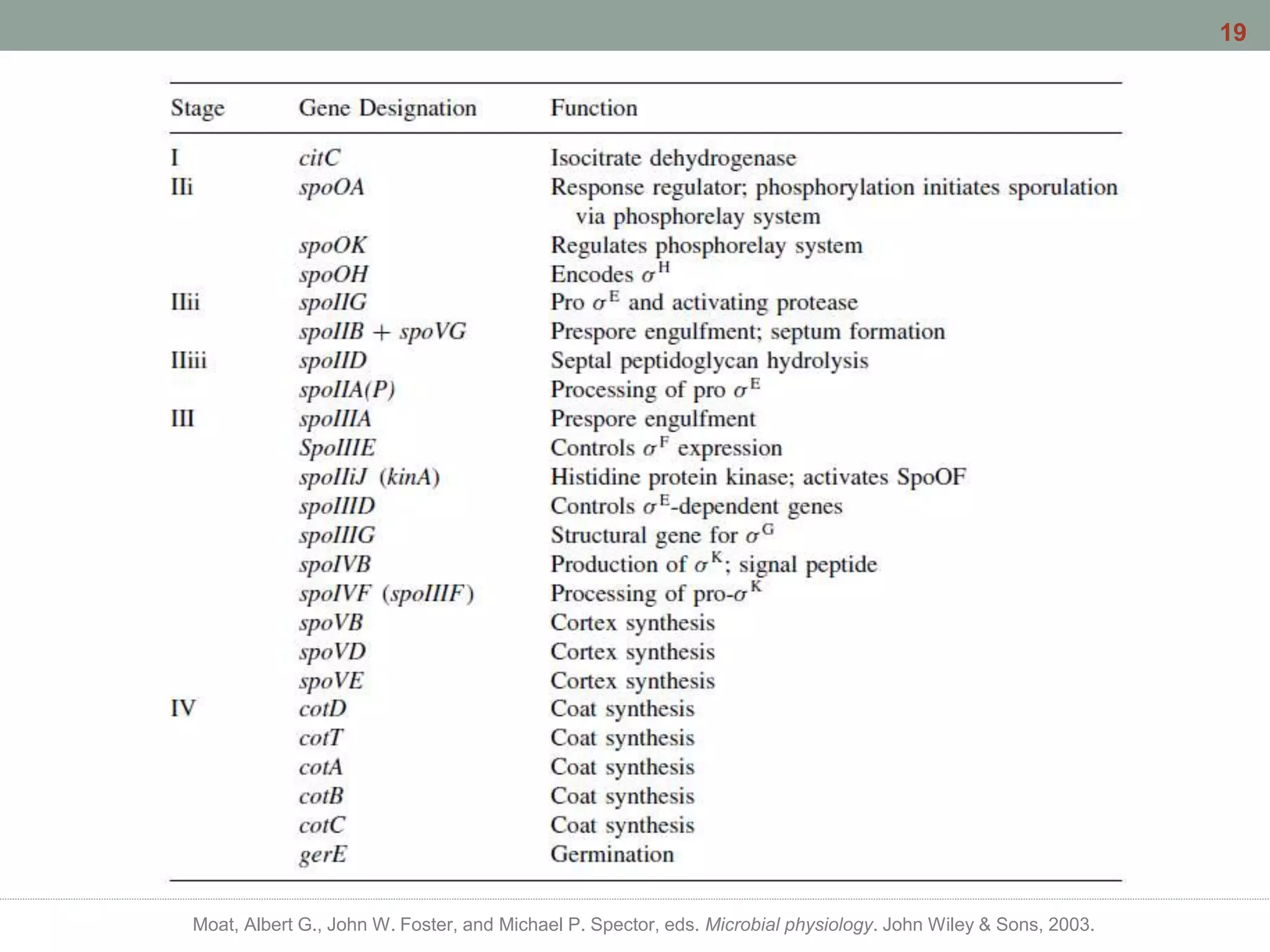
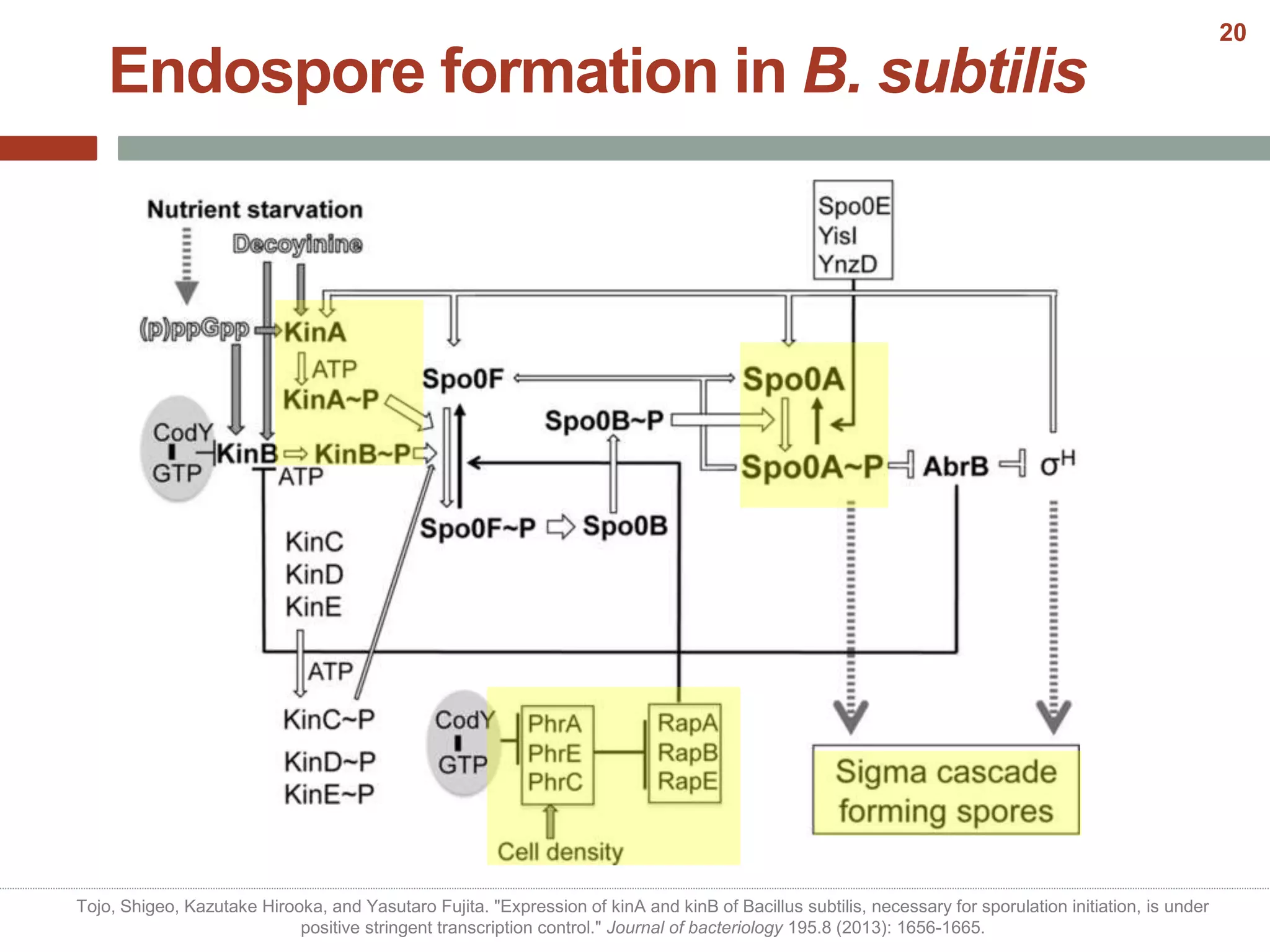
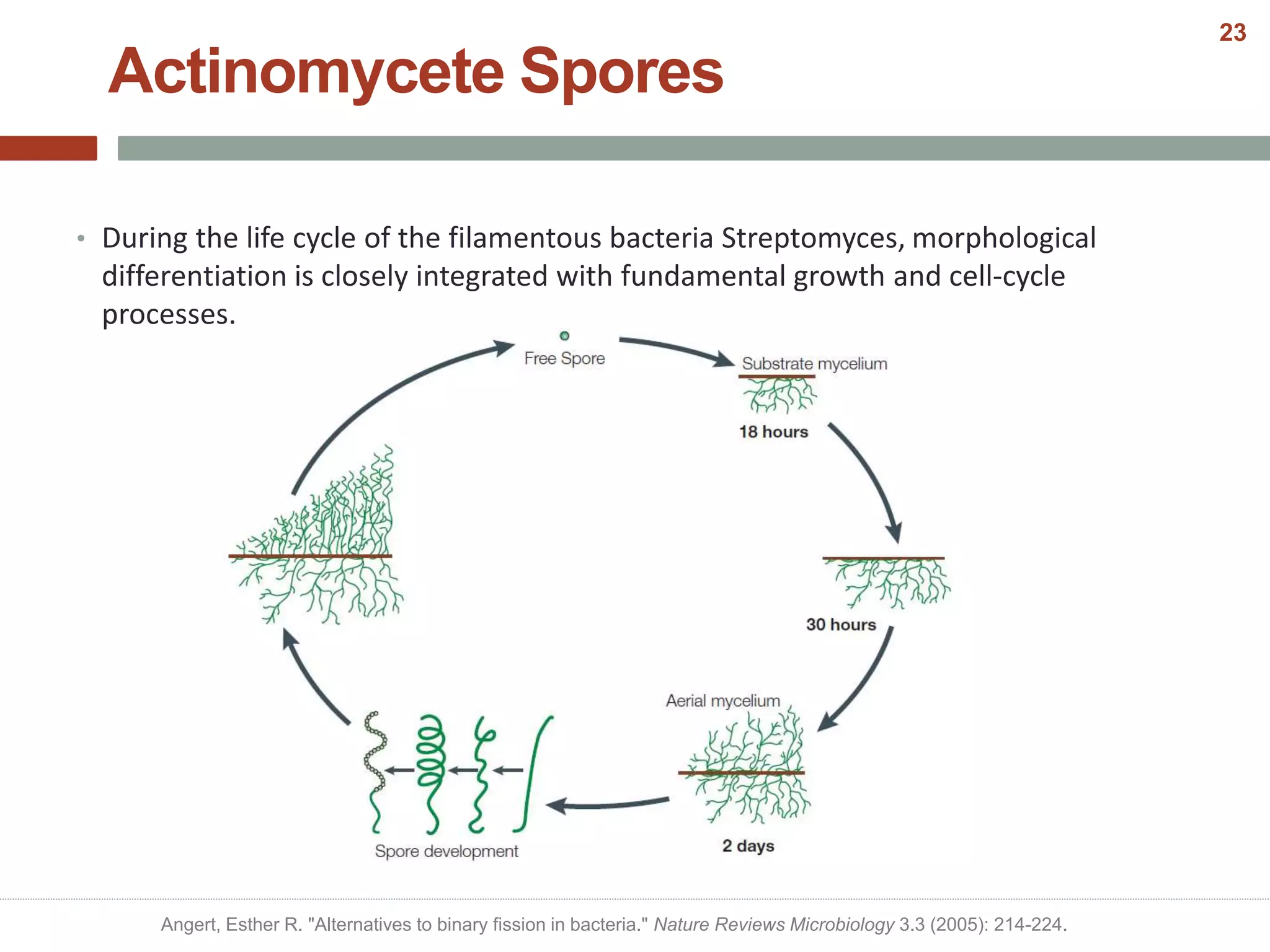
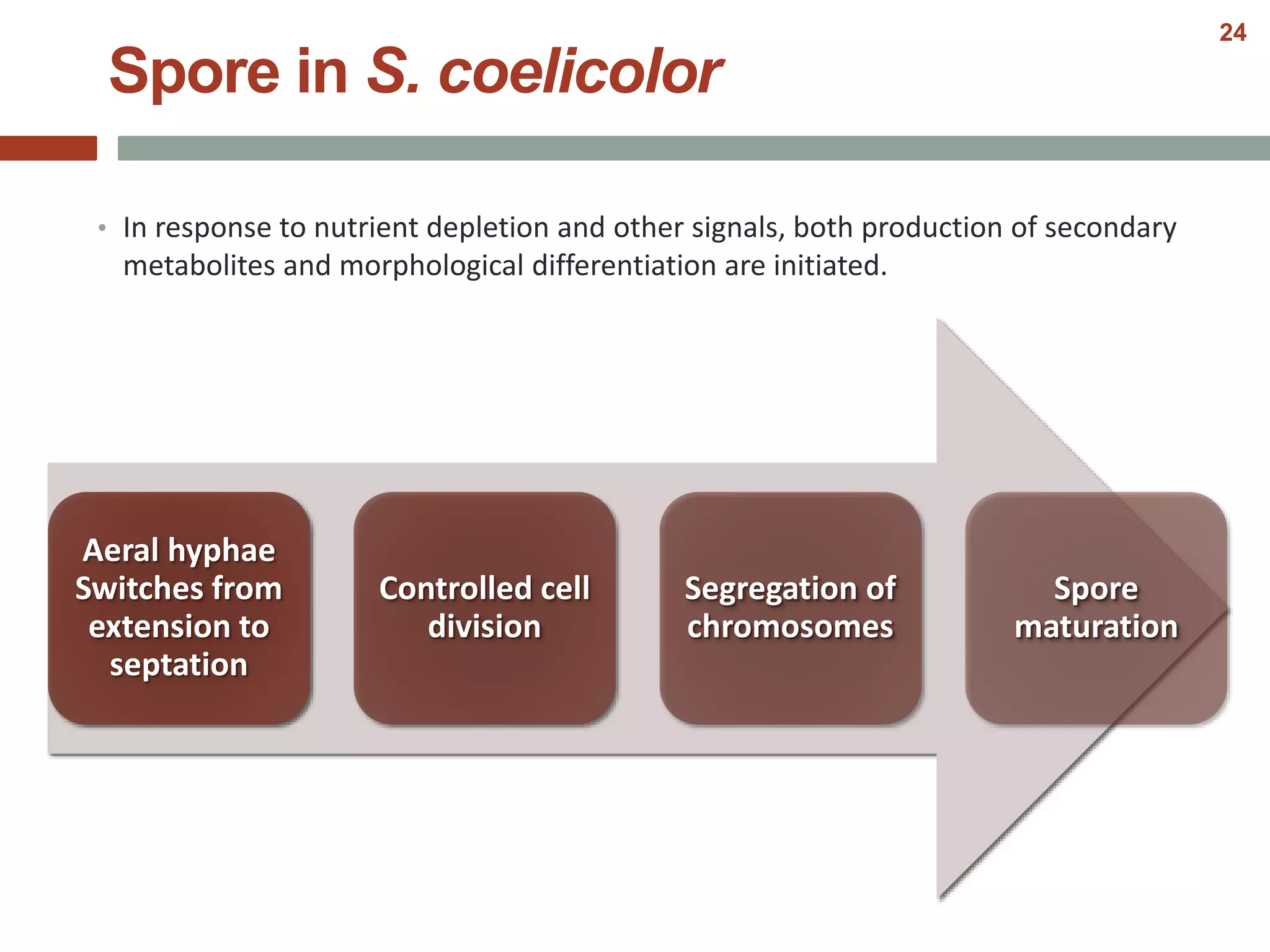
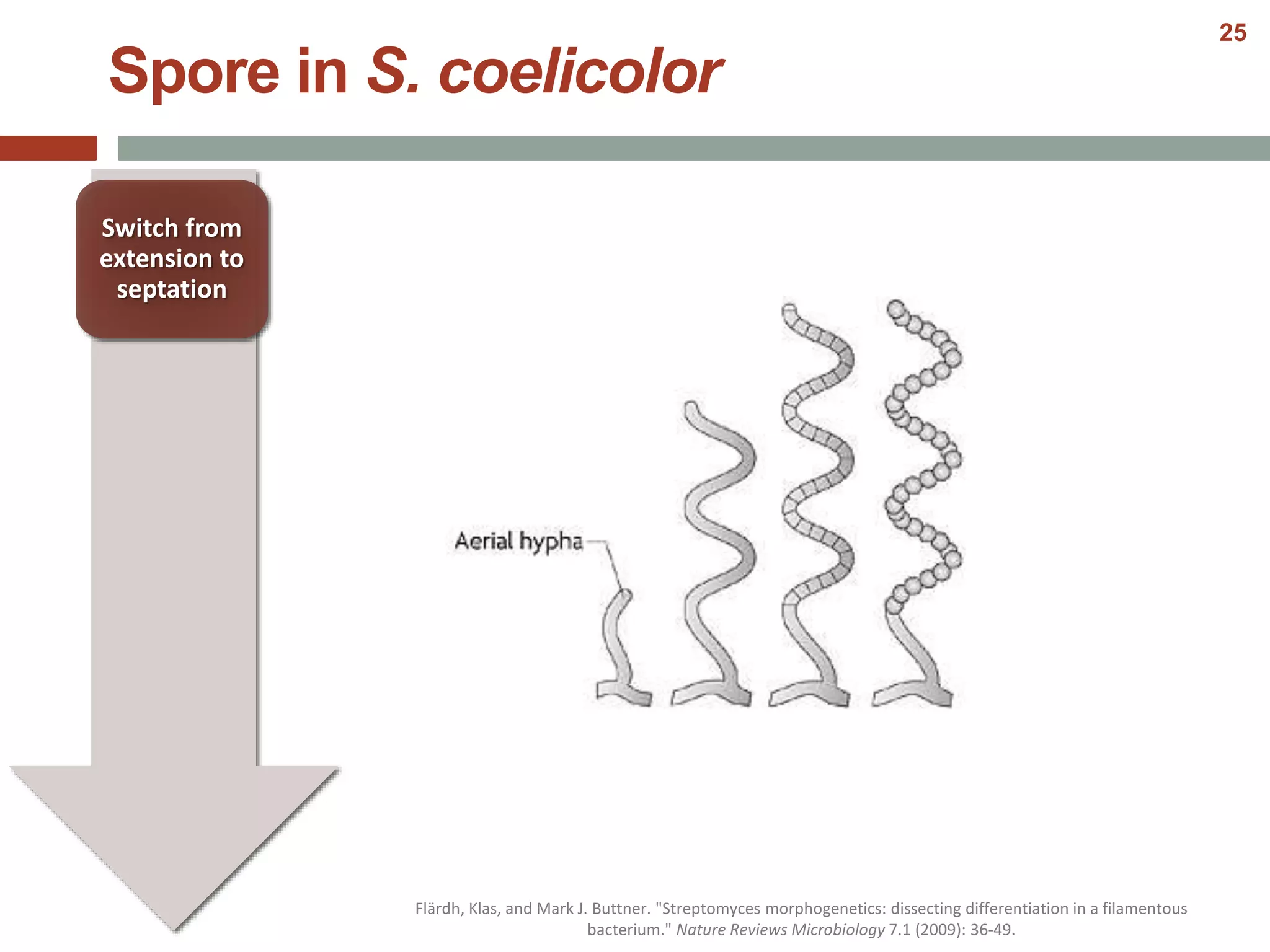
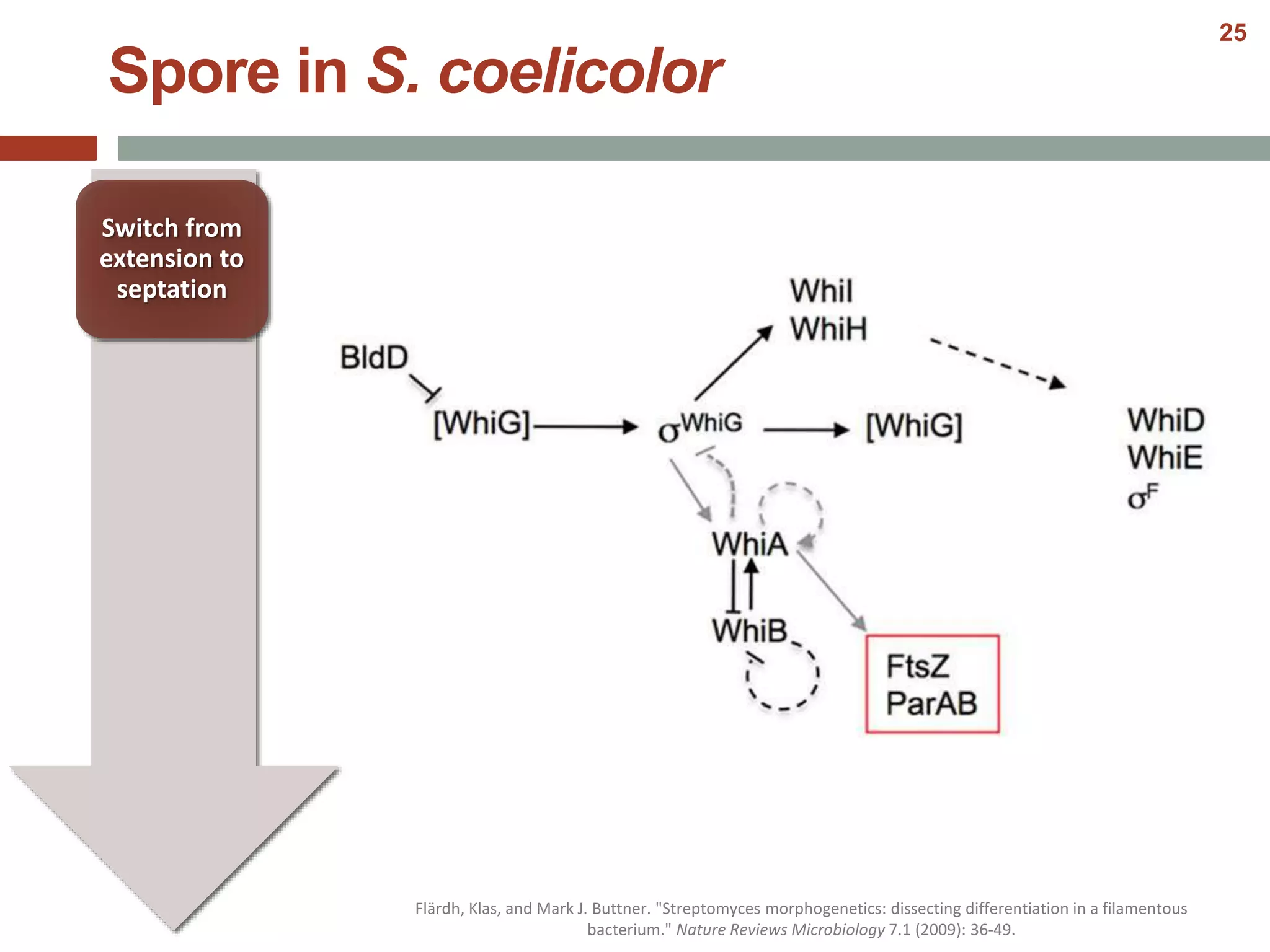
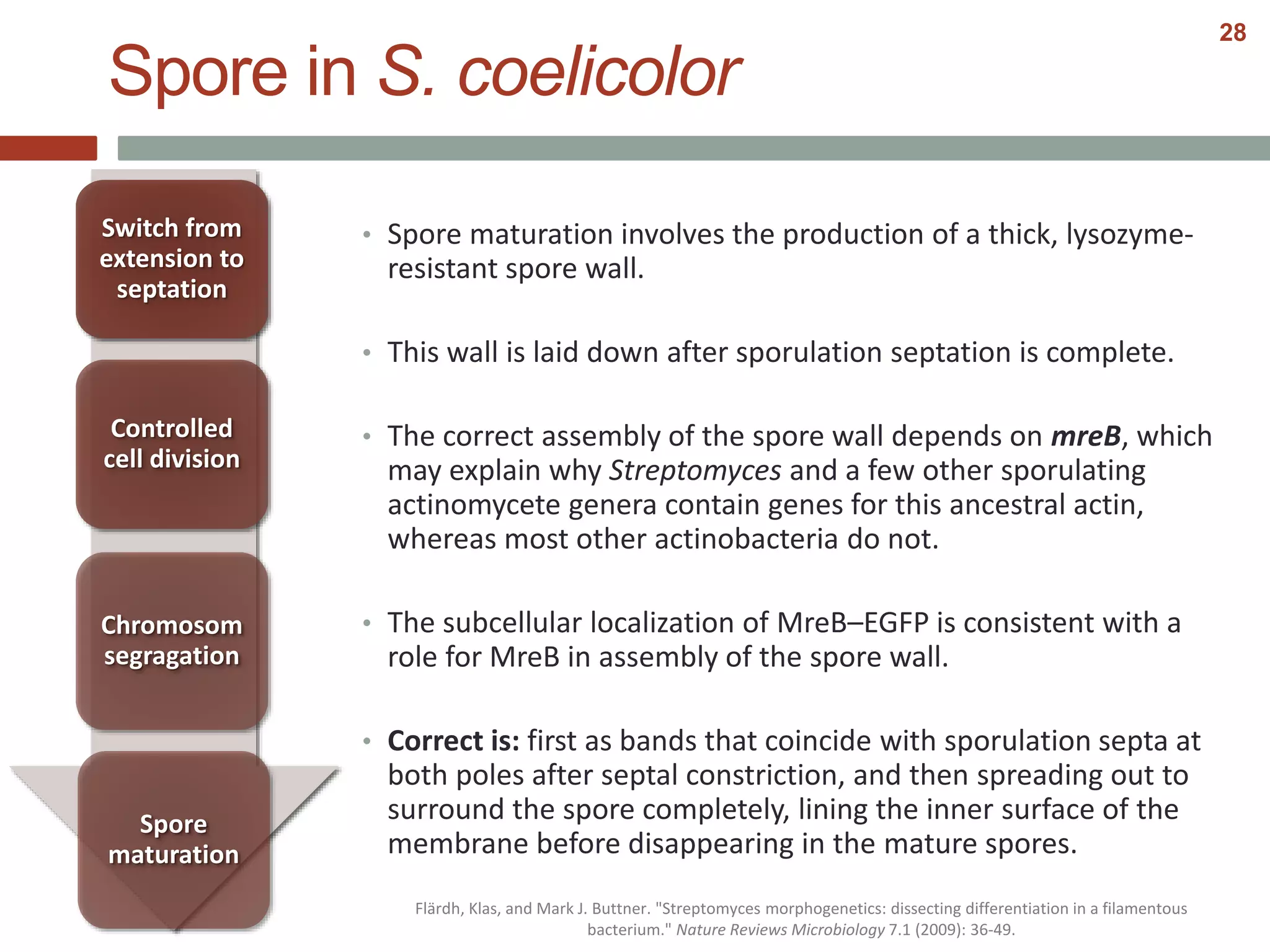
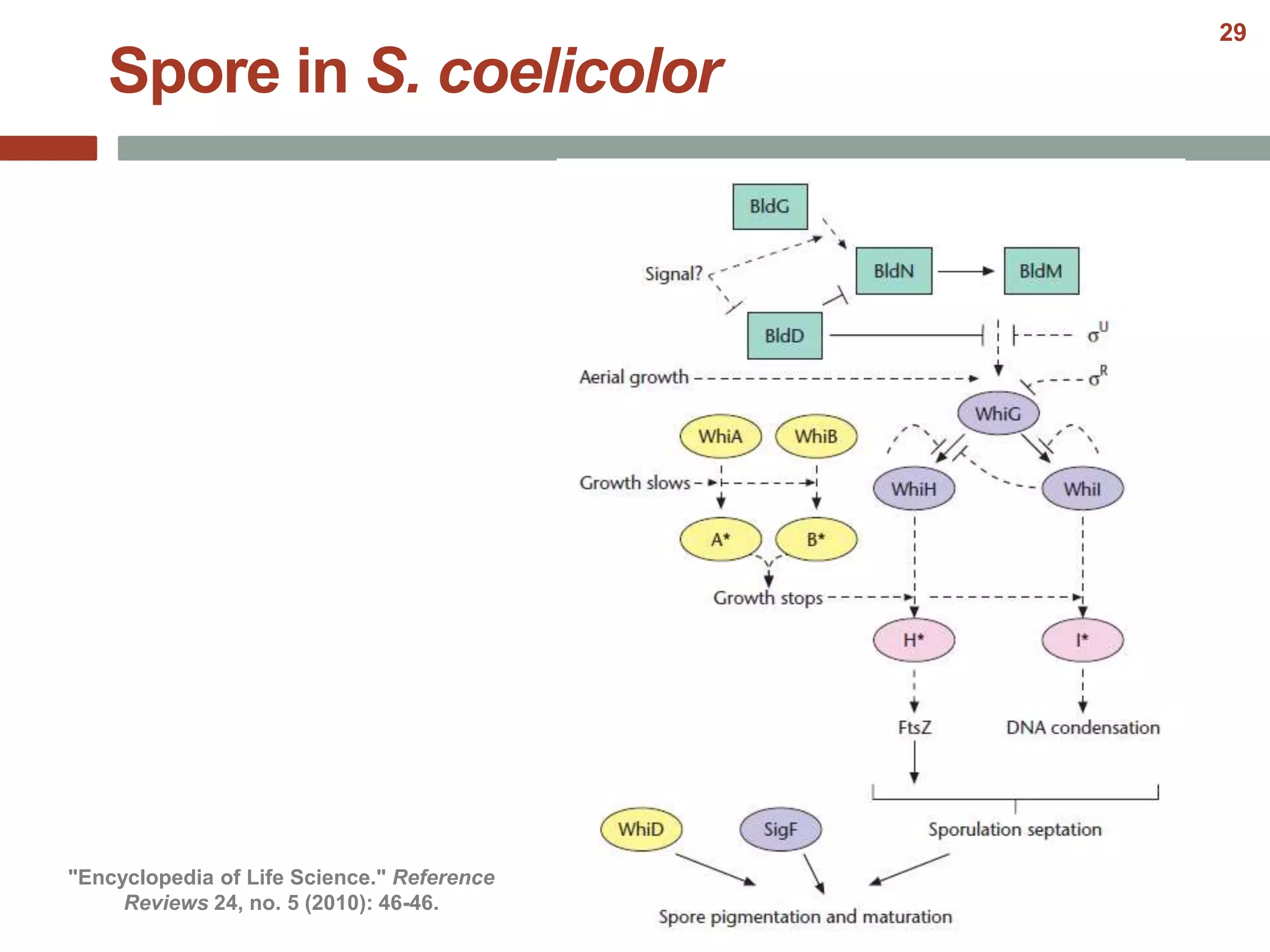
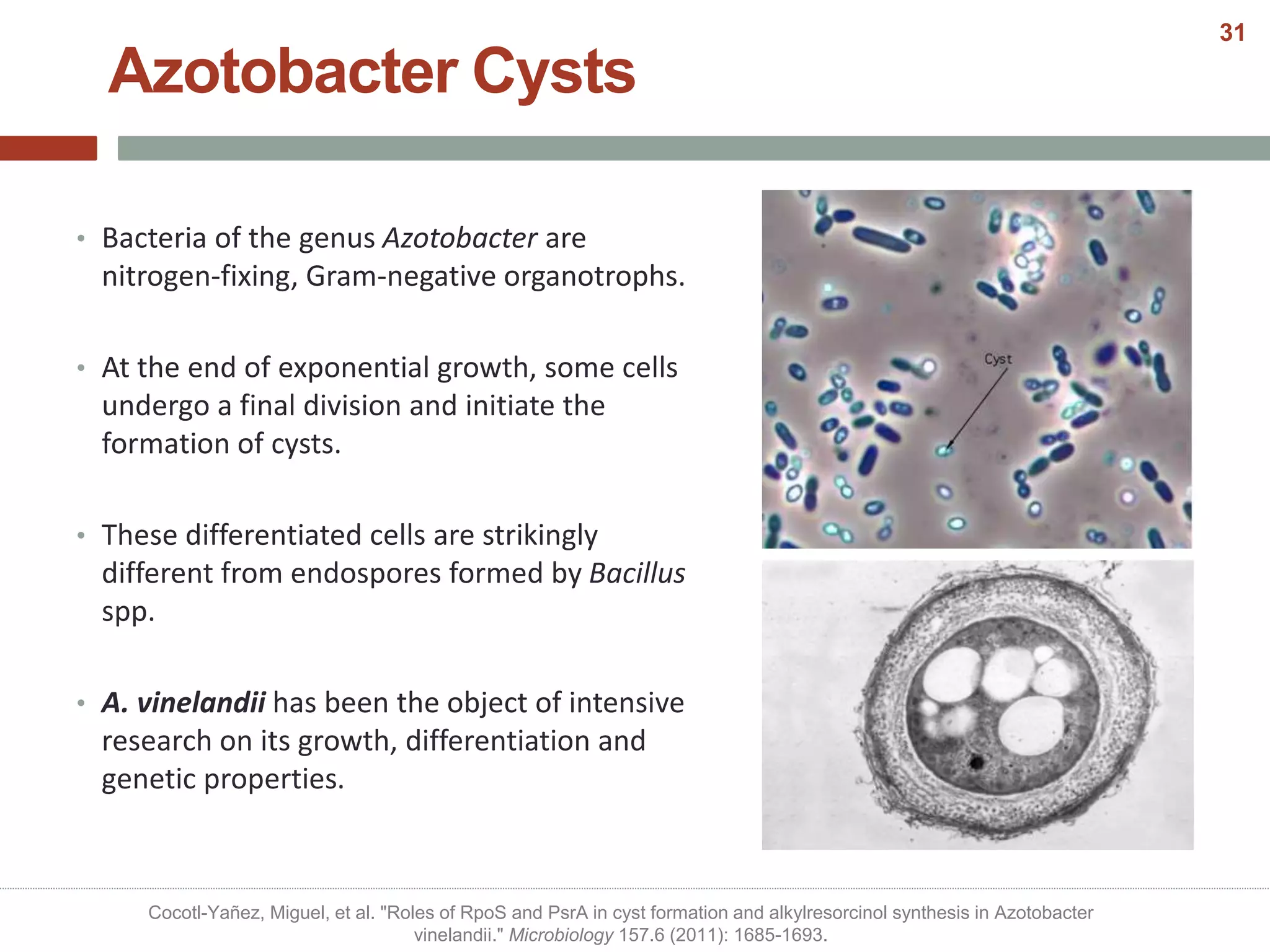
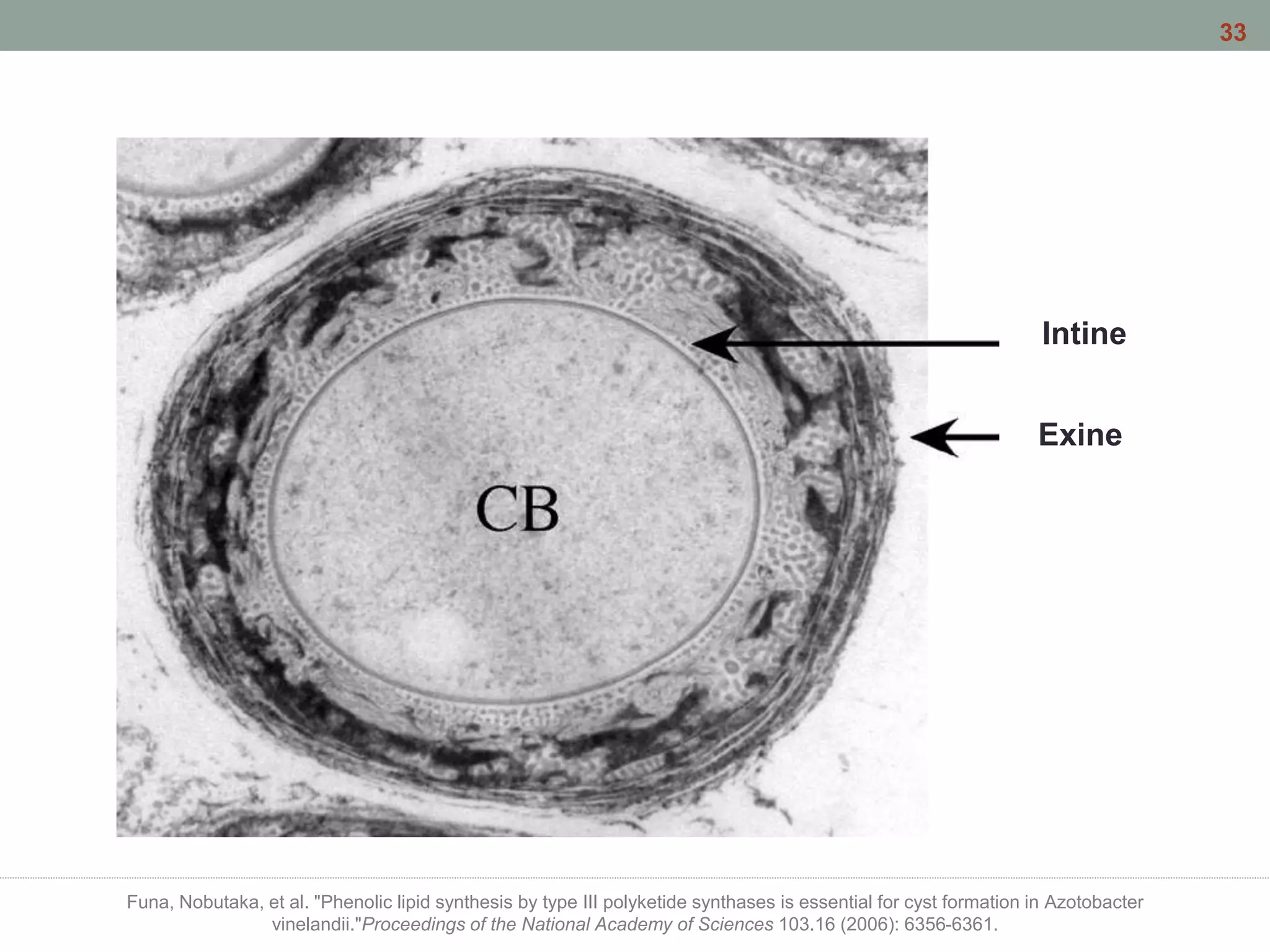
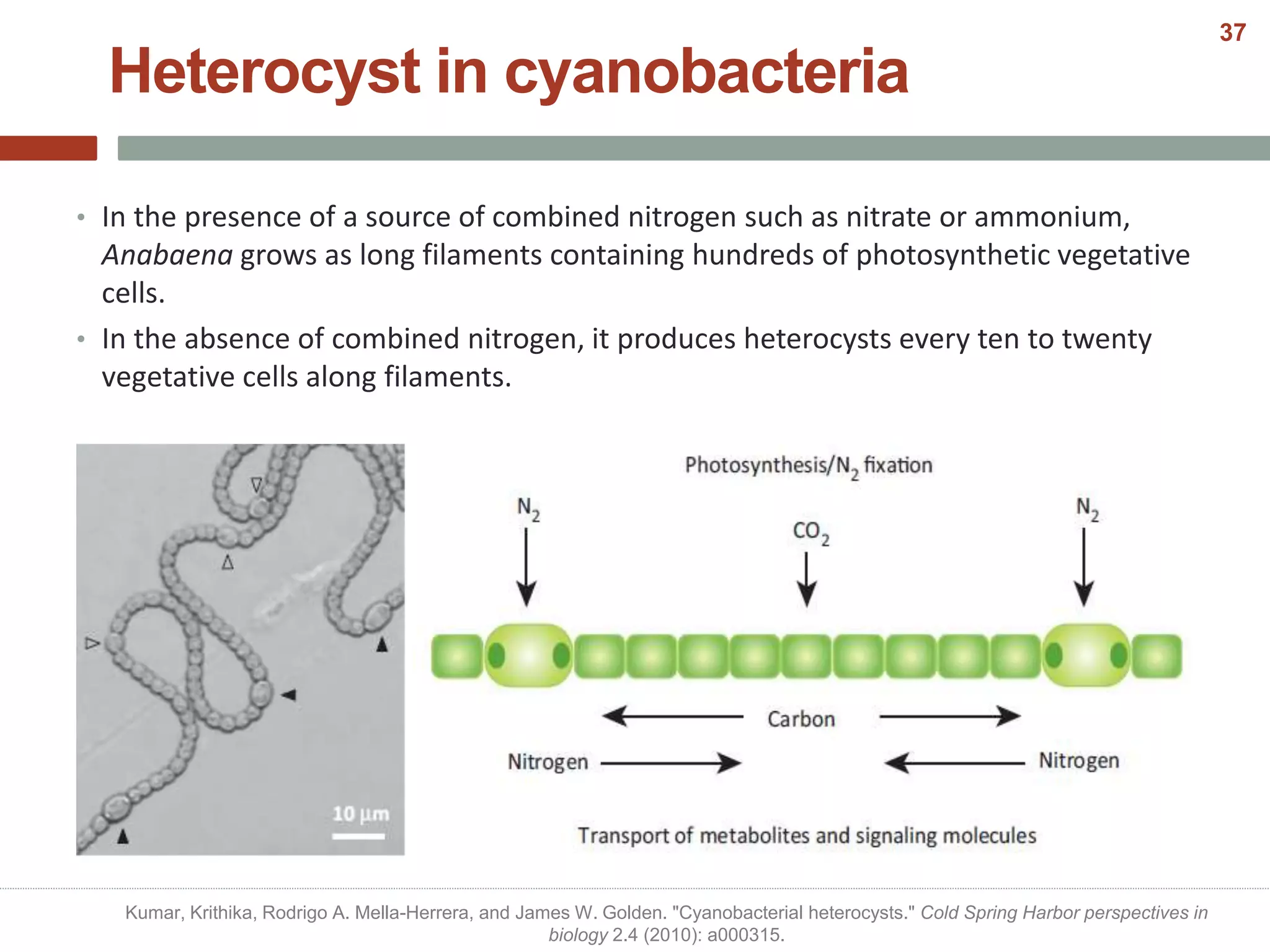
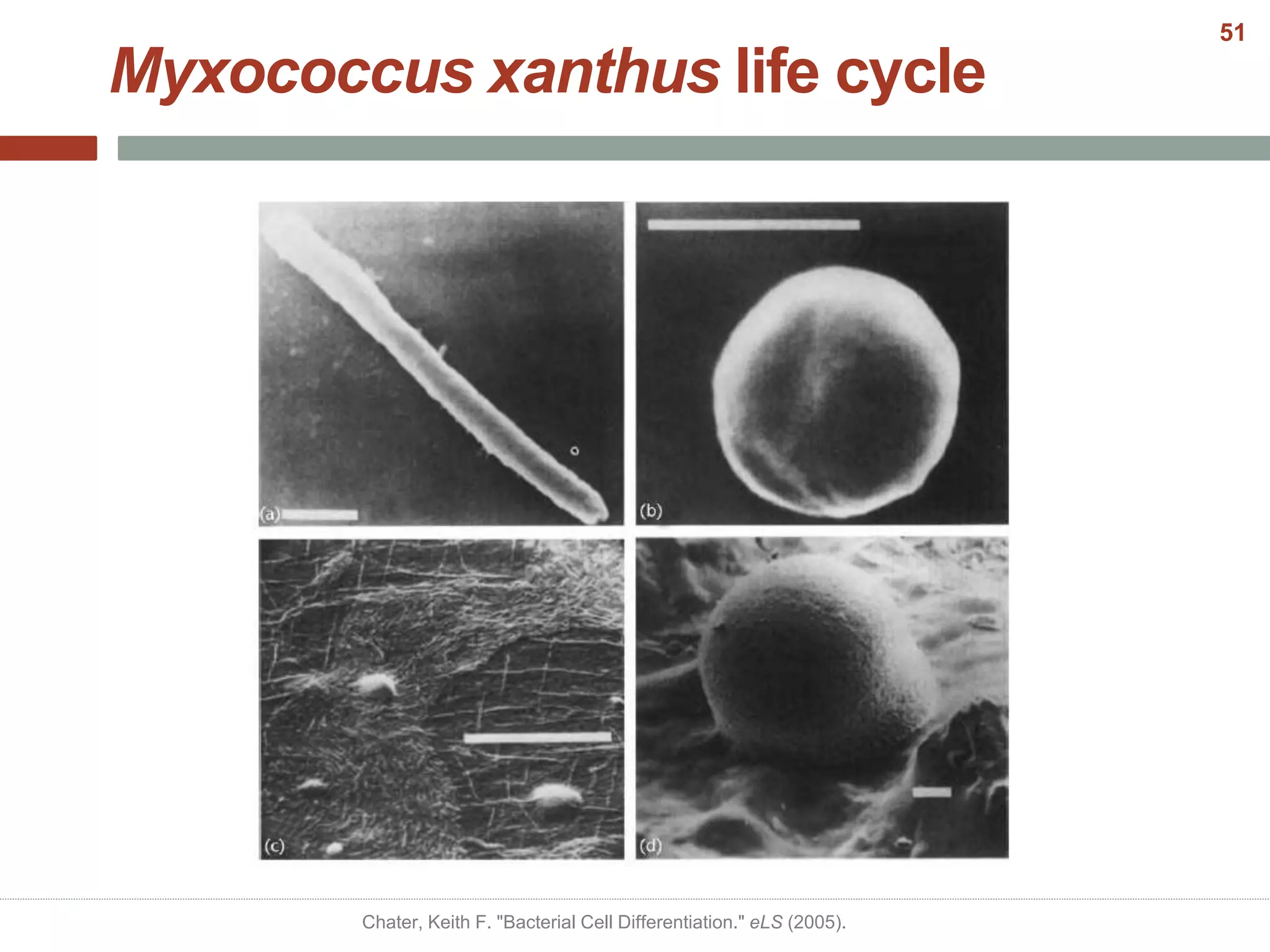
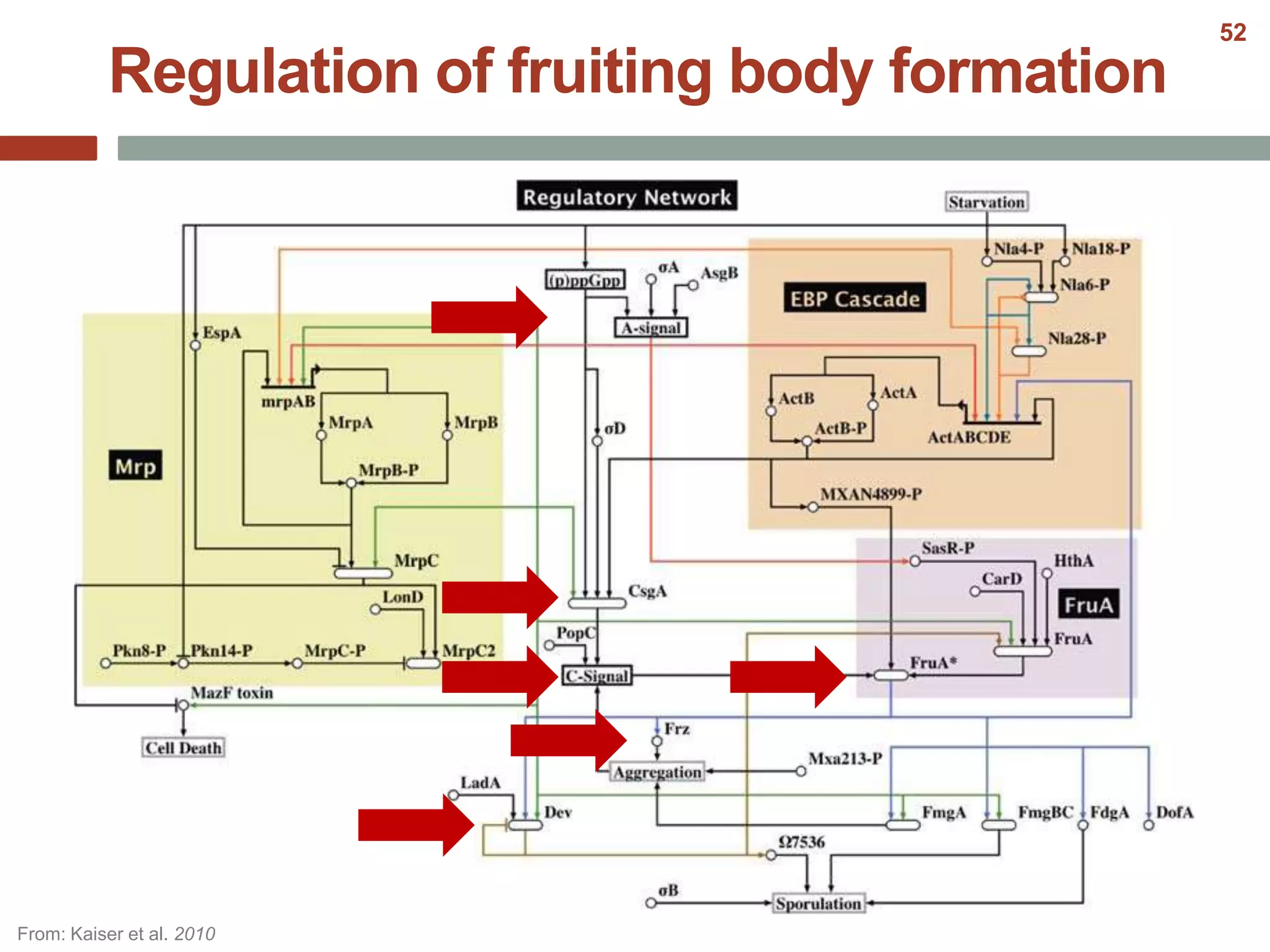
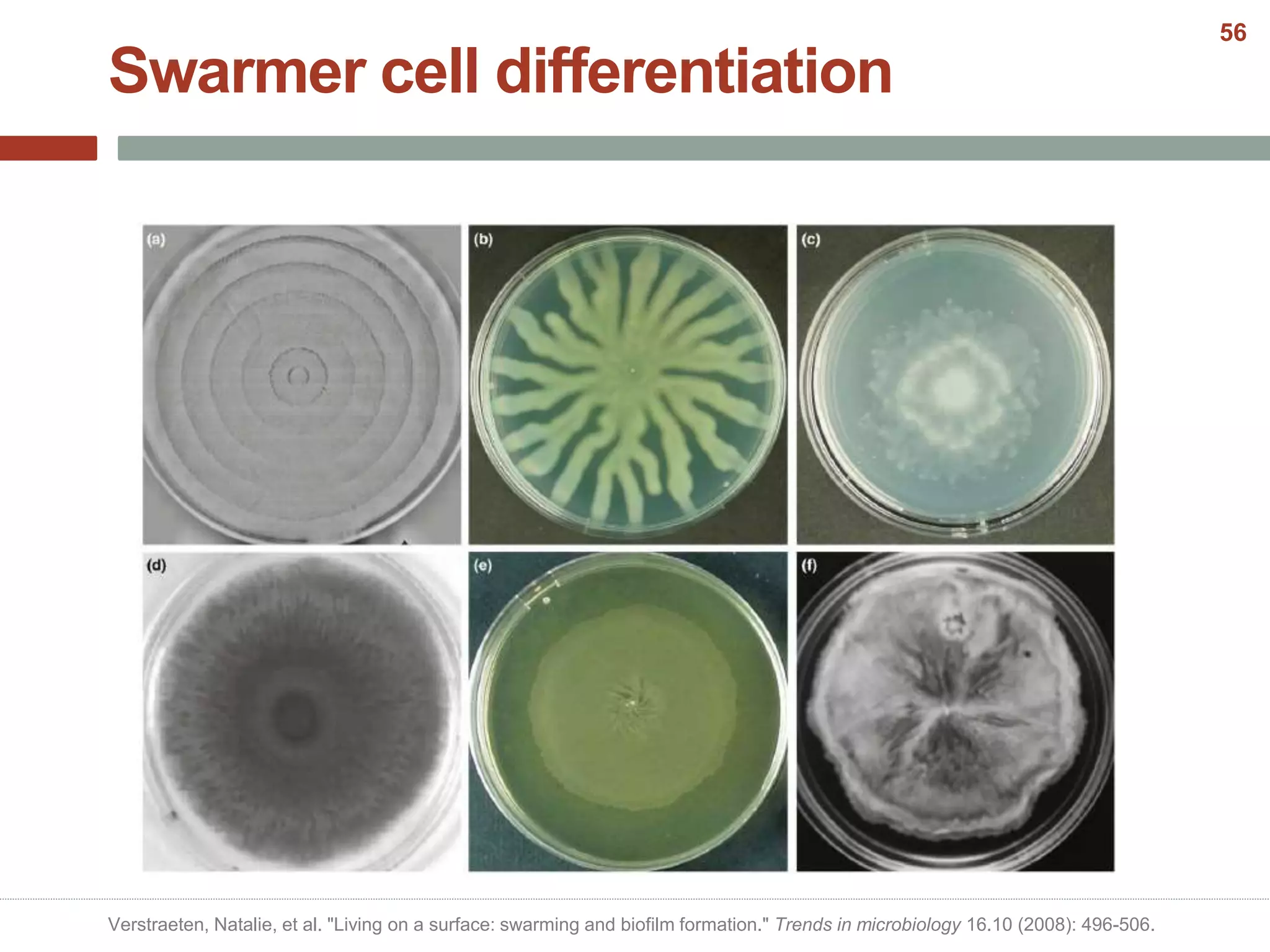
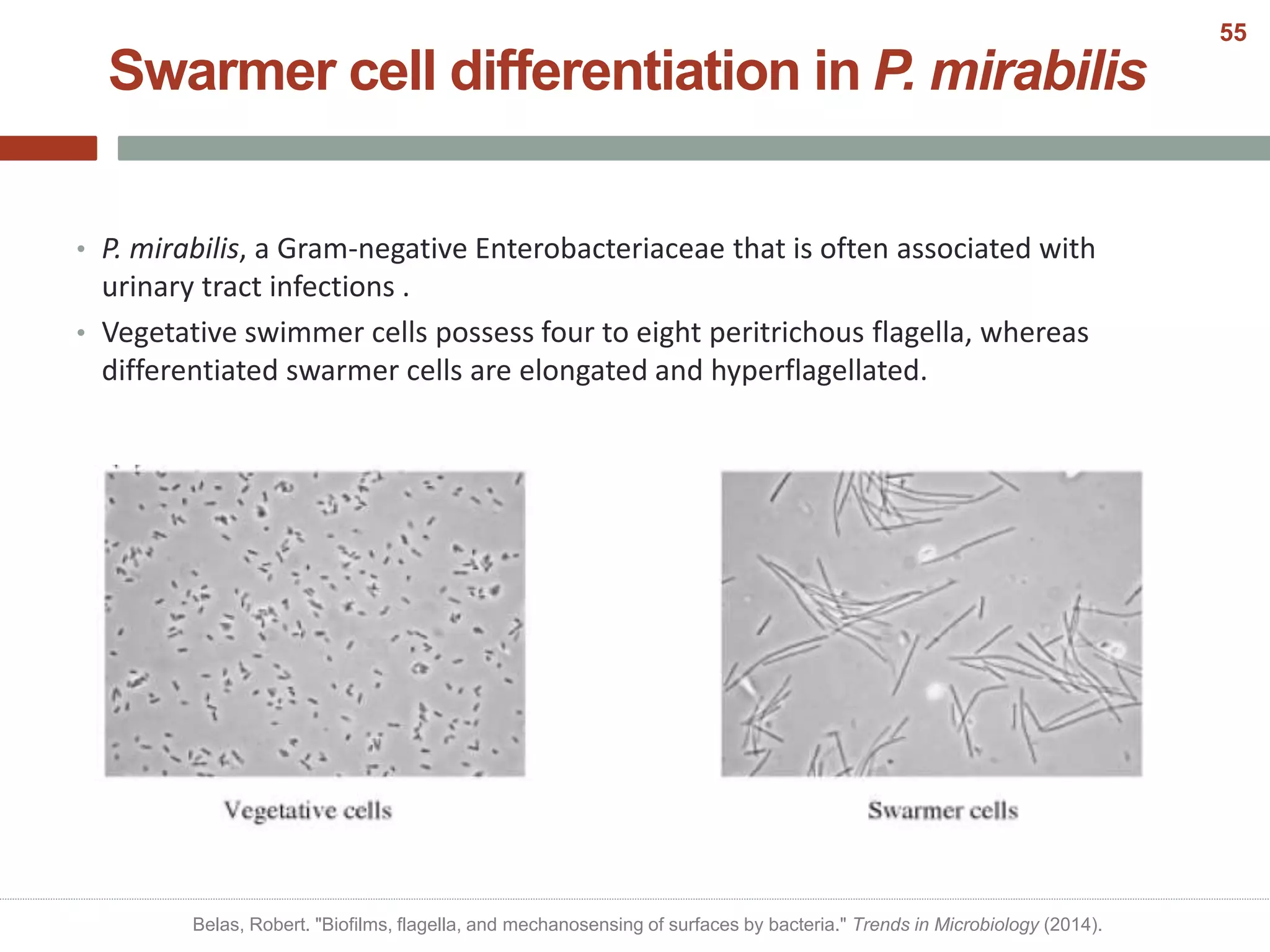
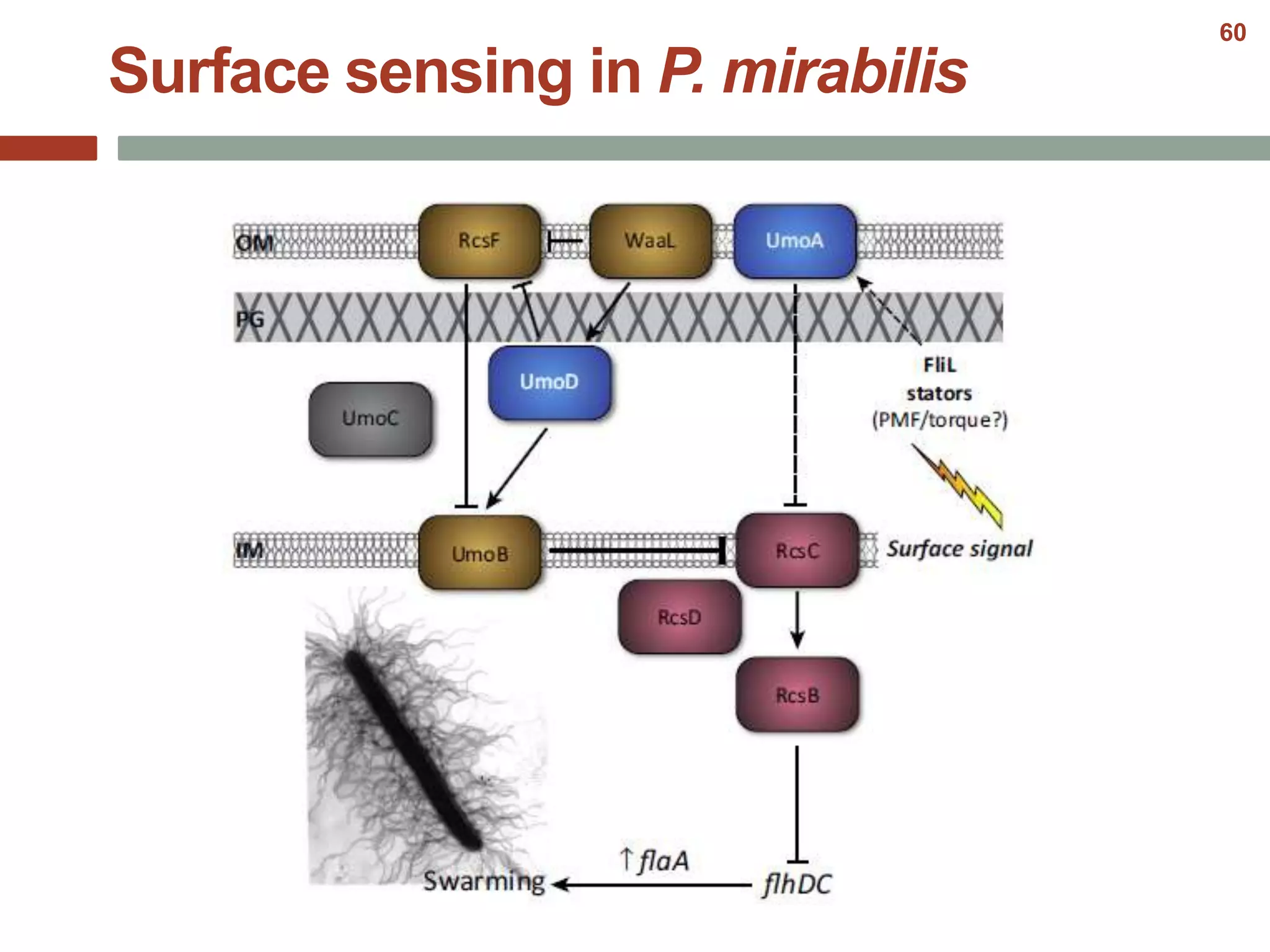
This document discusses bacterial cell differentiation, emphasizing the significance of morphological changes in response to environmental conditions and the complex regulatory pathways involved. It covers various differentiation examples including swarmer and stalked cell types in Caulobacter crescentus, endospore formation in Bacillus subtilis, and the formation of cysts and heterocysts in Azotobacter and cyanobacteria, respectively. The differentiation processes are crucial for survival, adaptation, and the reproductive strategies of bacteria.