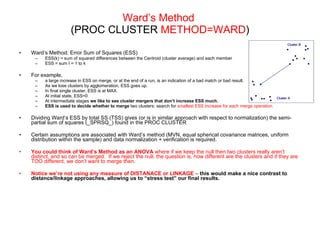
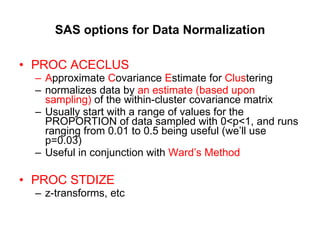
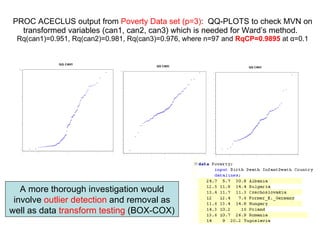
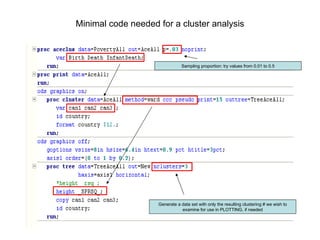
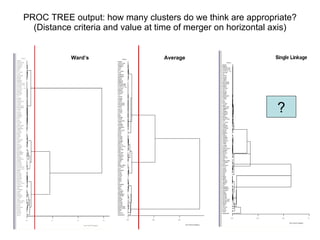
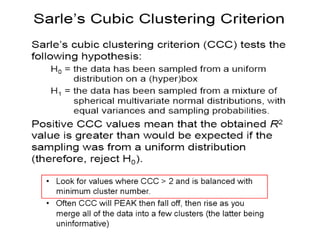
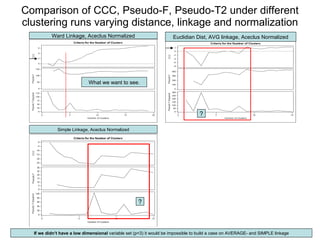
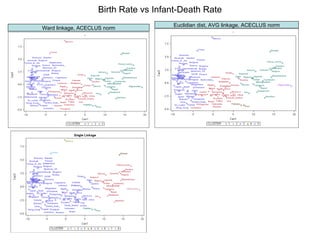
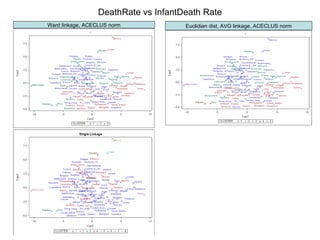
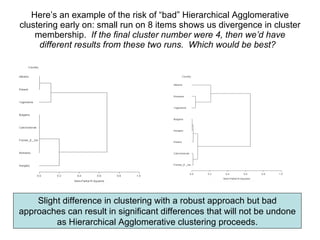
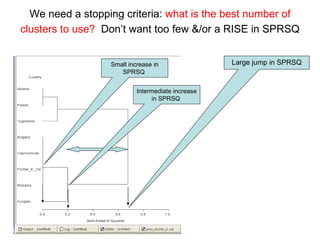
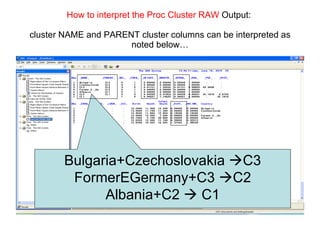
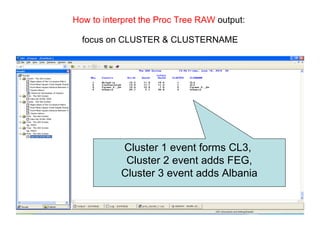
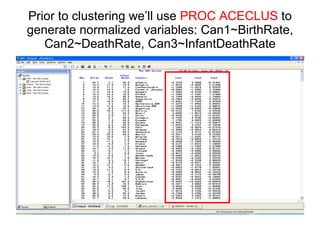
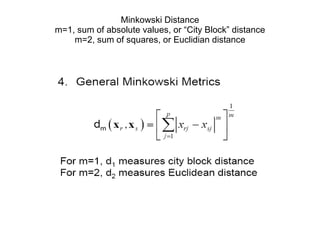
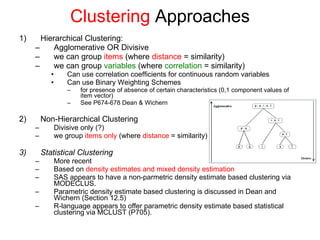
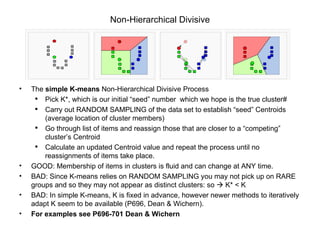
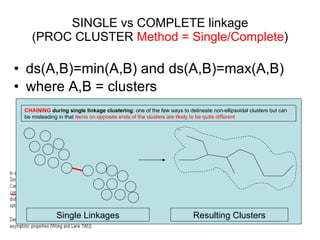
This document discusses different approaches to multivariate data analysis and clustering, including nearest neighbor methods, hierarchical clustering, and k-means clustering. It provides examples of using Ward's method, average linkage, and k-means clustering on poverty data to identify potential clusters of countries based on variables like birth rate, death rate, and infant mortality rate. Key lessons are that different linkage methods, distance measures, and data normalizations should be tested and that higher-dimensional data may require different variable spaces or transformations to identify meaningful clusters.





![AVERAGE linkage (PROC CLUSTER Method = AVERAGE) Σ [MxN d(a i ,b i )]/ (MxN) As one would expect, less influenced by outliers than SINLGLE or COMPLETE [ d(A1,B1),d(A1,B2),d(A1,B3) d(A2,B1),d(A2,B2),d(A2,B3) d(A3,B1),d(A3,B2),d(A3,B3) ]/9](https://image.slidesharecdn.com/nearestneighborv31-12786220622471-phpapp01/85/Statistical-Clustering-9-320.jpg)