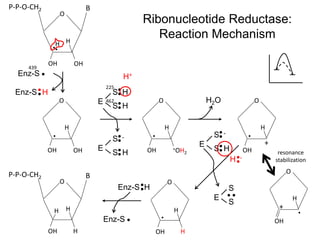
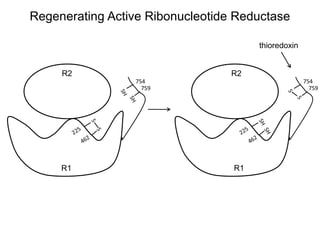
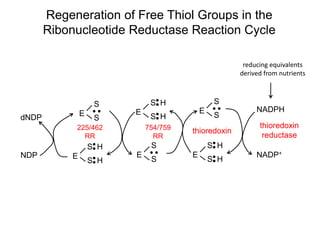
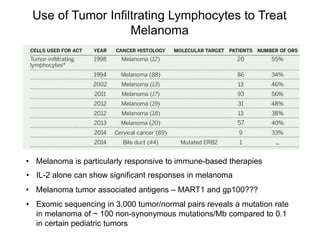
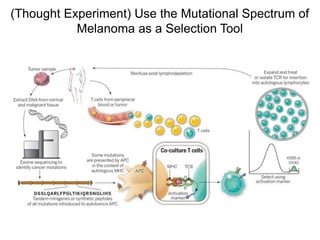
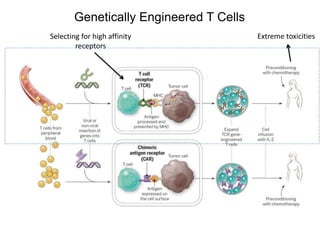
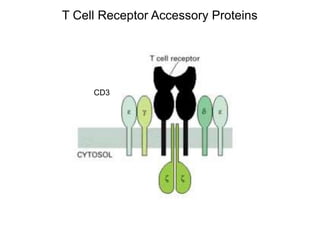
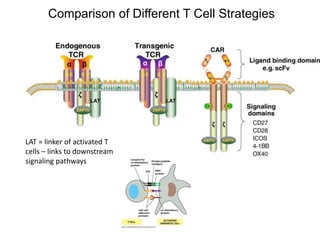
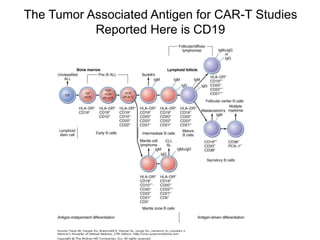
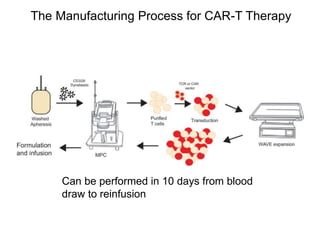
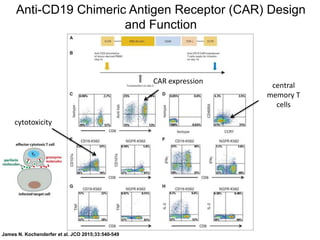
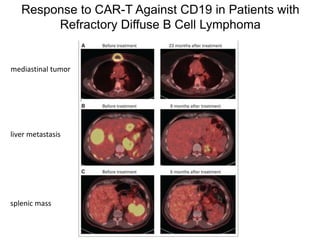
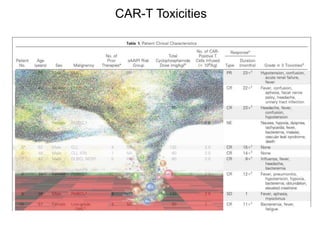
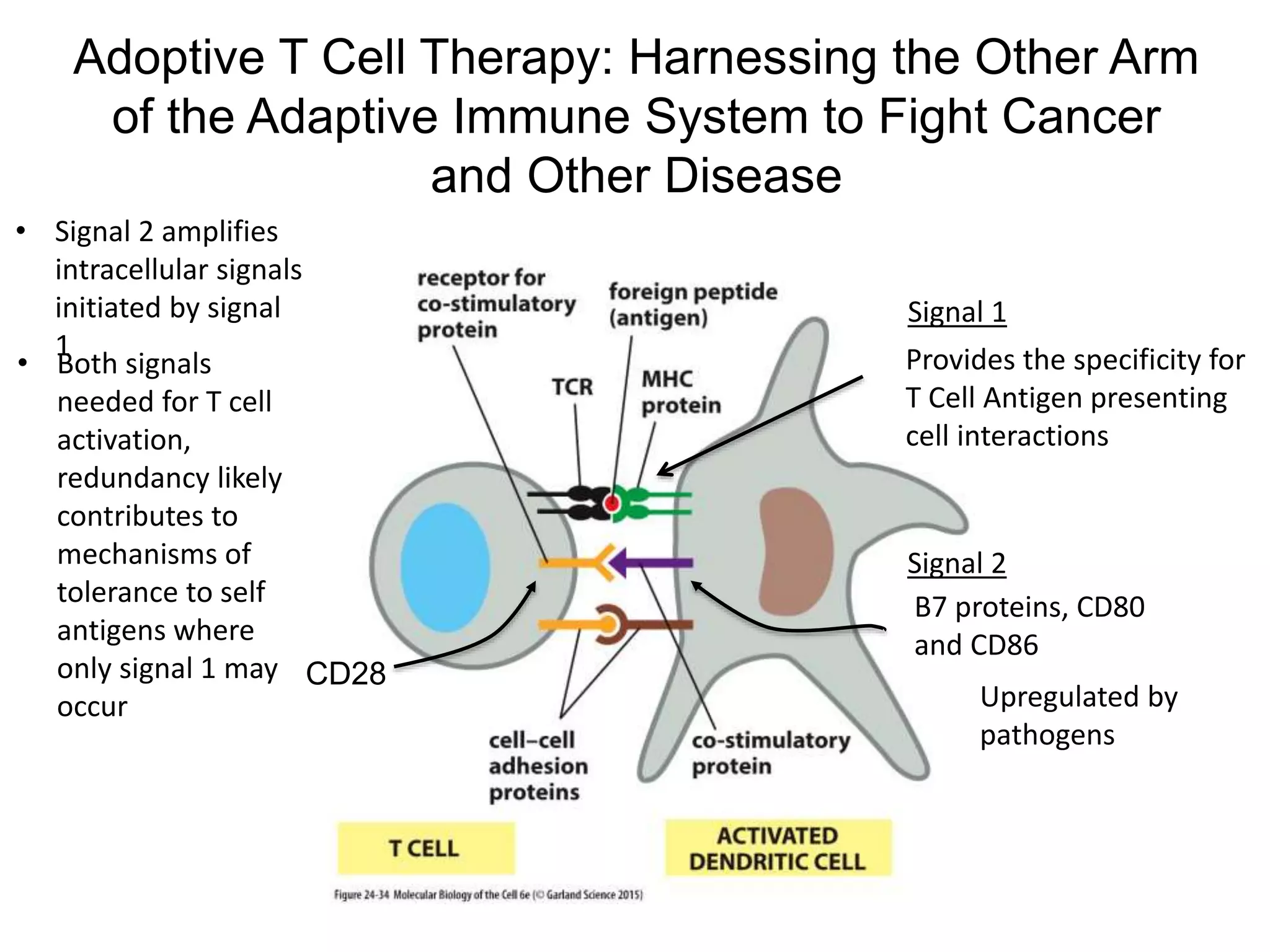
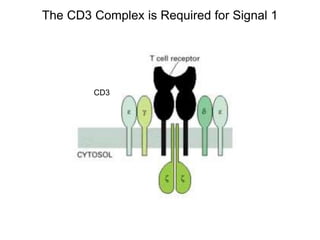
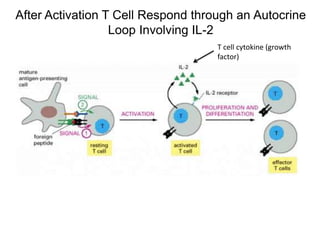
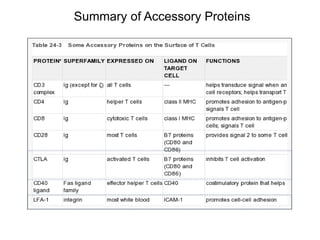
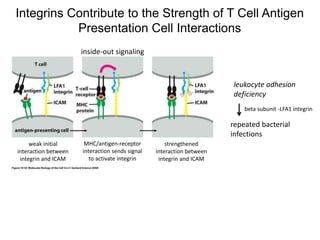
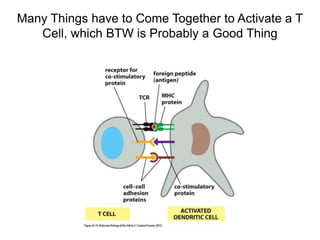
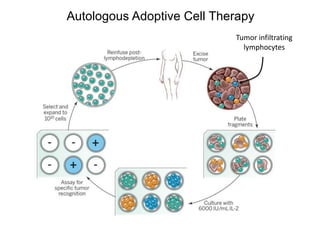
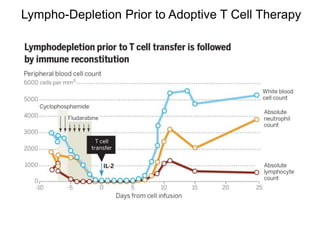
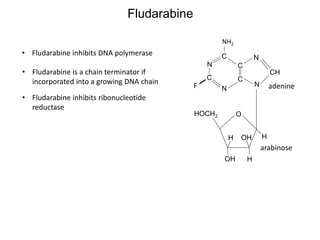
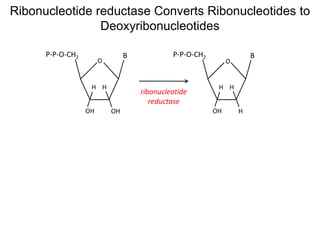
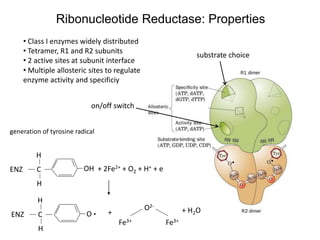
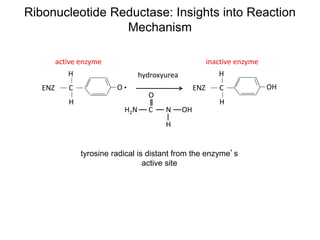
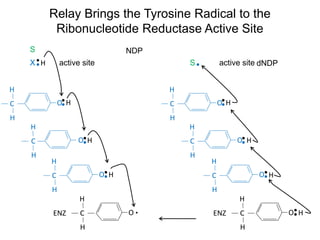
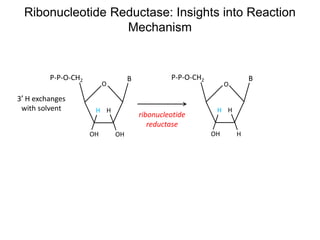
This document discusses adoptive T cell therapy and strategies to harness the adaptive immune system to fight cancer and other diseases. It provides an overview of T cell activation pathways and the role of accessory proteins like CD28 and CD3. It also summarizes methods to engineer T cells, including using tumor-infiltrating lymphocytes and genetically modifying T cells to express chimeric antigen receptors targeting cancers like CD19-positive leukemia. The document discusses approaches like lympho-depletion prior to therapy and highlights some toxicities seen with CAR-T therapy.
![freeenergy(G)
reaction progress
S
P
-ΔG (P-S)
activation energy
(uncatalyzed reaction)
activation energy
(catalyzed reaction)
energy state
of substrates
energy state
of products
T*
transition
state
S P
reaction spontaneous
as written
Enzyme Energetics
[ES]](https://image.slidesharecdn.com/lecture7adoptivetcelltherapy2-150619021051-lva1-app6892/85/adoptive-T-cell-therapy-15-320.jpg)