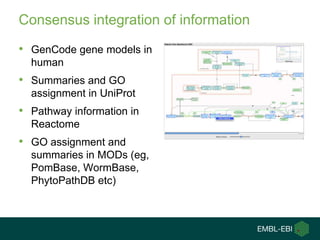
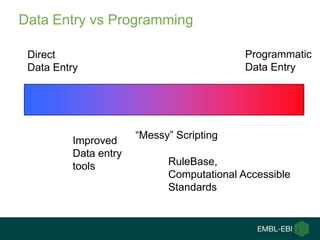
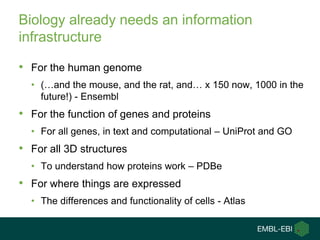
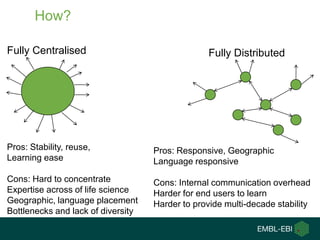
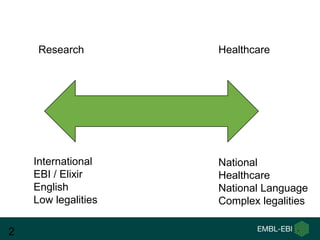
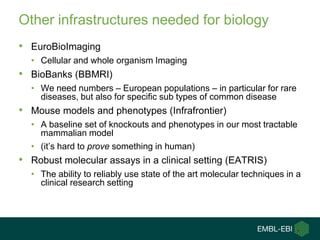
Ewan Birney summarizes the types of curation needed in molecular biology. There are five main types: experimental data entry, experimental metadata capture, consensus integration of information, knowledge frameworks, and knowledge management. Curation is critical for biology to integrate and organize the vast amounts of data being generated, but curation work is often overlooked despite its complexity. Large infrastructures are needed to support curation efforts and ensure biological data and knowledge can be reliably accessed and built upon over decades.