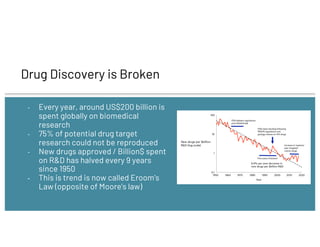
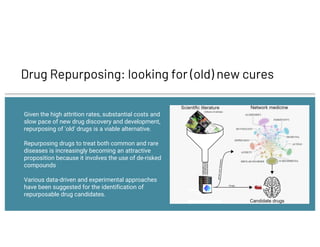
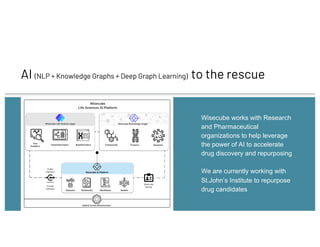
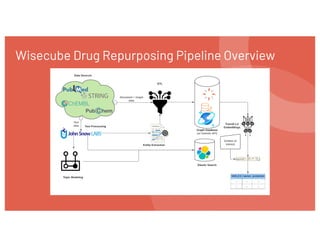
The document discusses drug repurposing using deep learning and knowledge graphs as a solution to the inefficiencies in drug discovery, highlighting the roles of data scientists Alex Thomas and Vishnu at Wisecube. It emphasizes the advantages of repurposing existing drugs for various diseases due to lower costs and de-risked compounds, while detailing a pipeline for building and utilizing a drug repurposing knowledge graph. The collaboration with St. John's Institute to repurpose drugs for Alzheimer's disease is also noted, showcasing the application of AI in accelerating research.


![Datasets: DRKG
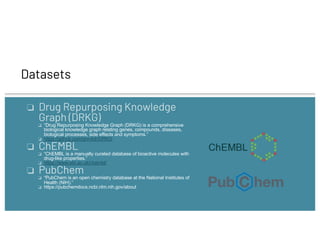
❏ DrugBank
❏ “DrugBank is a pharmaceutical knowledge base that is enabling major advances across the data-driven medicine
industry.”
❏ Link: https://go.drugbank.com/
❏ GNBR
❏ “A global network of biomedical relationships derived from text”
❏ https://zenodo.org/record/1134693#.WqQe1GbVSL9
❏ Hetionet
❏ “Hetionet is an integrative network of biomedical knowledge assembled from 29 different databases of genes,
compounds, diseases, and more.”
❏ https://het.io/
❏ StringDB
❏ “STRING is a database of known and predicted protein-protein interactions.”
❏ https://string-db.org/cgi/about
❏ IntAct
❏ “IntAct provides a freely available, open source database system and analysis tools for molecular interaction data.
“
❏ https://www.ebi.ac.uk/intact/
❏ DGIdb
❏ “[I]nformation on drug-gene interactions and the druggable genome, mined from over thirty trusted sources.”
❏ https://www.dgidb.org/
HETIONET](https://image.slidesharecdn.com/27alexthomas-210614165035/85/Drug-Repurposing-using-Deep-Learning-on-Knowledge-Graphs-9-320.jpg)


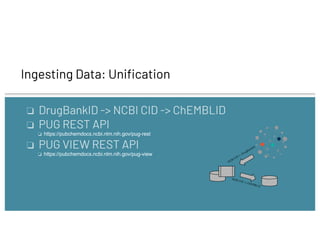
![Ingesting Data: Loading
❏ Ingest into Graph DB
❏ Neptune
❏ CosmosDB
❏ Any Graph DB which supports Gremlin
❏ Graph DB vs Triple Store
❏ Most open data is in RDF triples formats (RDF/XML, Turtle,
N-Triples)
❏ Modern Graph Dbs are faster than Triple Stores
@prefix sio: <http://semanticscience.org/resource/> .
@prefix compound: <http://rdf.ncbi.nlm.nih.gov/pubchem/compound/> .
@prefix descriptor: <http://rdf.ncbi.nlm.nih.gov/pubchem/descriptor/> .
compound:CID400516 sio:has-attribute
descriptor:CID400516_Isomeric_SMILES ,
descriptor:CID400516_Isotope_Atom_Count ,
descriptor:CID400516_Molecular_Formula ,
descriptor:CID400516_Molecular_Weight ,
descriptor:CID400516_Mono_Isotopic_Weight ,
descriptor:CID400516_Non-hydrogen_Atom_Count ,
~id ~label articles:String[] source_ids:String[] name:String SMILES:String
8647 COMPOUND 13961;... CHEMBL1200689 Nitric oxide [N]=O
344 COMPOUND 268975;... CHEMBL142438 Nitrogen N#N
18030 COMPOUND 10081;... CHEMBL925 TYROSINE N[C@@H](Cc1ccc(O)cc1)C(=O)O
1534 COMPOUND 211538;... CHEMBL1616046
HYPOCHLOROUS
ACID
OCl
18800 COMPOUND 13464;... CHEMBL978 Methacholine CC(=O)OC(C)C[N+](C)(C)C
26747 COMPOUND 226005;.... CHEMBL863 Cysteine N[C@@H](CS)C(=O)O](https://image.slidesharecdn.com/27alexthomas-210614165035/85/Drug-Repurposing-using-Deep-Learning-on-Knowledge-Graphs-13-320.jpg)