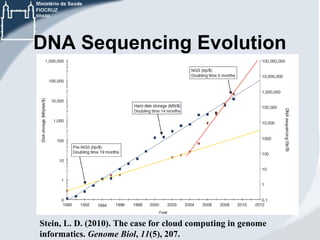
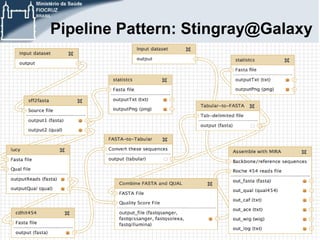
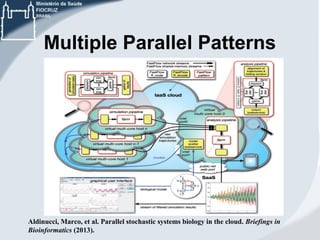
This document discusses the use of cloud computing technologies for genomic big data analysis. It begins by defining big data and describing the exponential growth of genomic data. It then discusses how cloud computing provides flexibility, scalability, and accessibility for genomic data processing through virtualization and large computing clusters. Specific technologies enabled for the cloud that help with genomic analysis are described, such as Hadoop, MapReduce, and genomic analysis tools adapted for these frameworks. The document concludes by discussing challenges remaining around data transfer speeds and the need for cloud application expertise, but also describes how platforms like Galaxy Cloudman and Cloudgene allow genomic analysis in the cloud without programming expertise.