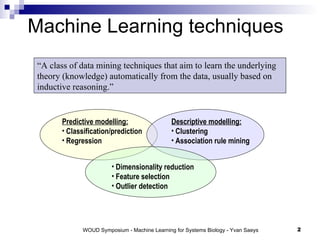
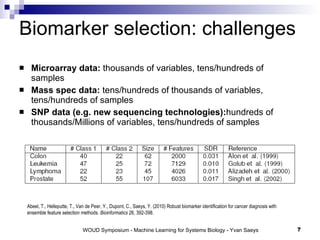
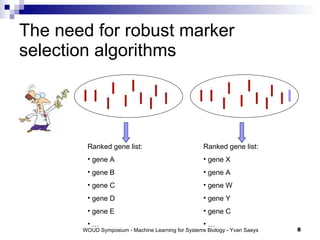
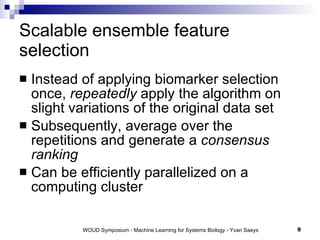
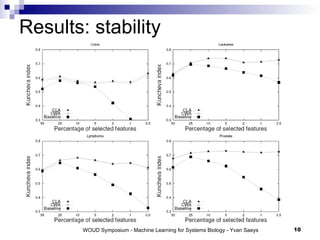
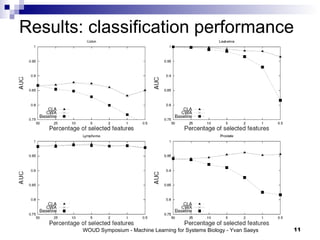
The document discusses large-scale machine learning challenges in systems biology, emphasizing the need for robust methodologies and data integration in predictive modeling. It highlights case studies on robust biomarker discovery, automated literature screening, and network inference, showcasing the importance of ensemble methods and high-performance computing. The findings suggest that incorporating model robustness as an evaluation criterion is essential for scalable learning models.
![Large scale machine learning challenges for systems biology Yvan Saeys Bioinformatics and Evolutionary Genomics (BEG) Department of Plant Systems Biology, VIB/UGent [email_address]](https://image.slidesharecdn.com/06yvan-110929012334-phpapp01/75/Large-scale-machine-learning-challenges-for-systems-biology-1-2048.jpg)



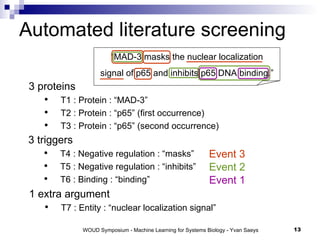
![Current state-of-the-art Extraction of specific biological relationships Potential for automatic summarization of articles Current performance [BioNLP Shared Task]](https://image.slidesharecdn.com/06yvan-110929012334-phpapp01/85/Large-scale-machine-learning-challenges-for-systems-biology-14-320.jpg)
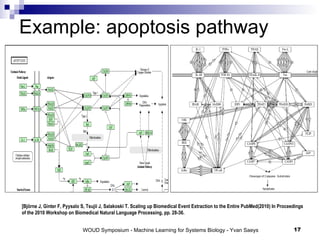
![From text mining to integrated networks [Saeys, Y., Van Landeghem, S., Van de Peer, Y. (2010) Event based text mining for integrated network construction. Journal of Machine Learning Research, Workshop and Conference proceedings 8, 112-121.] Binding/unspecied Regulation Phosphorylation Transcription Positive Regulation Negative Regulation](https://image.slidesharecdn.com/06yvan-110929012334-phpapp01/85/Large-scale-machine-learning-challenges-for-systems-biology-15-320.jpg)