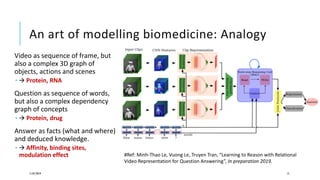
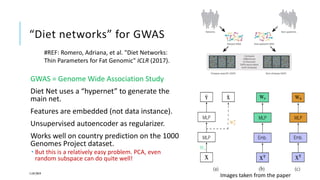
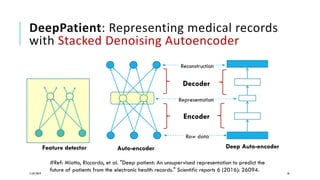
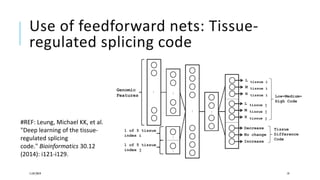
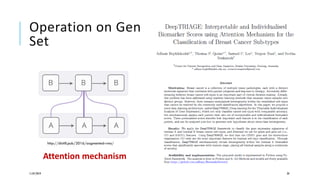
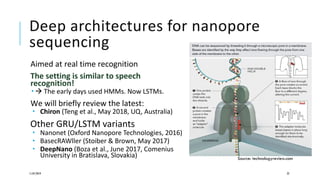
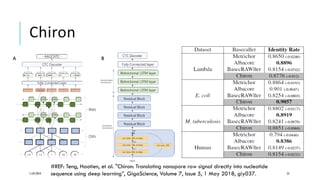
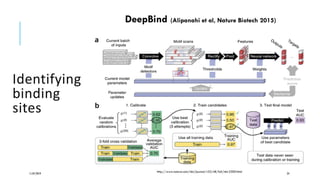
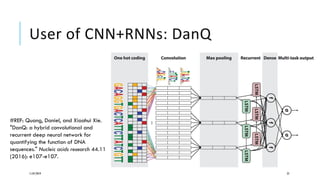
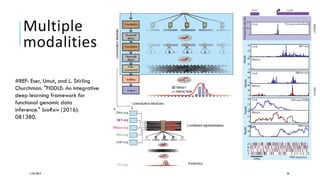
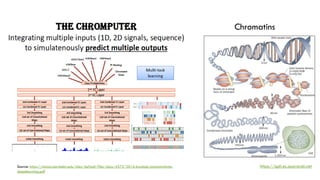
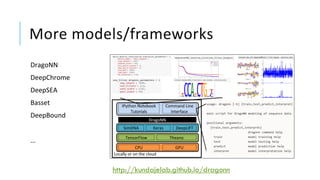
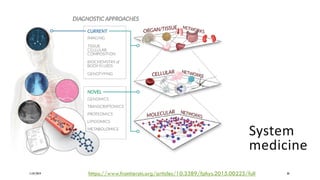
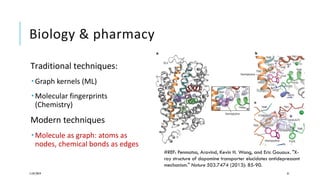
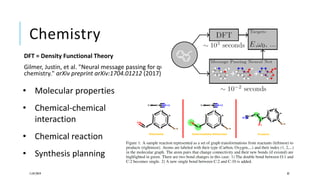
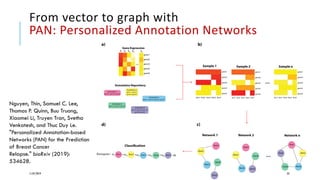
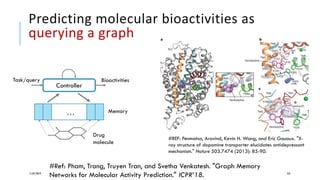
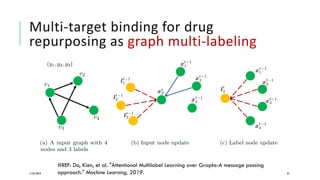
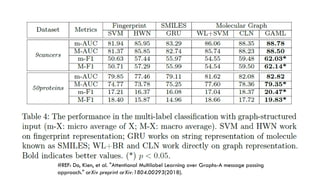
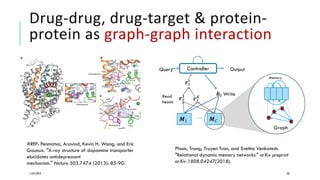
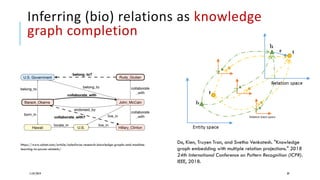
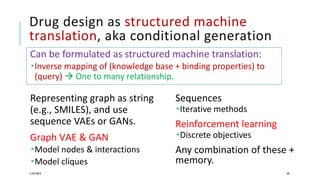
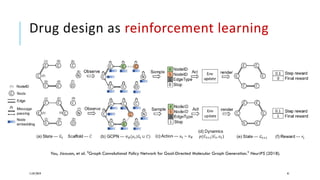
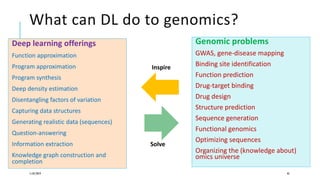
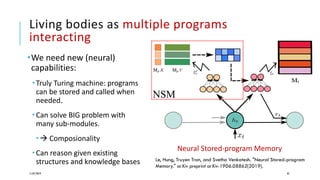
The document discusses the intersection of deep learning and biomedicine, highlighting recent advances in AI for drug design, genomic research, and health informatics. It emphasizes the potential impact of deep learning techniques on addressing complex biomedical challenges, such as high-dimensional data and privacy concerns. Various machine learning workshops and conferences focused on healthcare are mentioned, indicating an active research landscape devoted to improving healthcare through AI.

![Recent AI/ML/KDD activities
Conference on Machine Learning for Healthcare (MLHC), 2019
ICML/IJCAI/AAAI (2019)
Health Intelligence
Workshop on Computational Biology
Knowledge Discovery in Healthcare III: Towards Learning Healthcare Systems (KDH)
KDD/SDM/ICDM (2018-2019)
Health Day at KDD’18
epiDAMIK: Epidemiology meets Data Mining and Knowledge discovery
17th International Workshop on Data Mining in Bioinformatics
Workshop on Data Mining in Bioinformatics (BIOKDD 2019)
[DsHealth 2019] 2019 KDD workshop on Applied data science in Healthcare: bridging the gap
between data and knowledge
11/07/2019 5](https://image.slidesharecdn.com/seaml19-191027004530/85/Deep-learning-for-biomedicine-5-320.jpg)