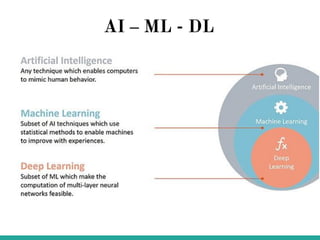
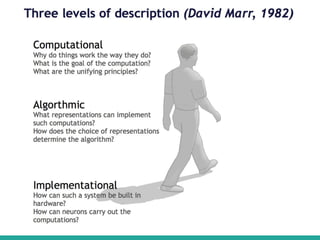
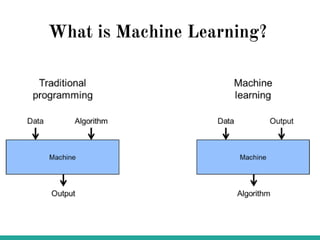
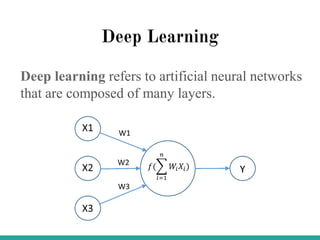
Machine learning has many applications and opportunities in biology, though also faces challenges. It can be used for tasks like disease detection from medical images. Deep learning models like convolutional neural networks have achieved performance exceeding human experts in detecting pneumonia from chest X-rays. Frameworks like DeepChem apply deep learning to problems in drug discovery, while platforms like Open Targets integrate data on drug targets and their relationships to diseases. Overall, machine learning shows promise for advancing biological research, though developing expertise through learning resources and implementing models to solve real-world problems is important.