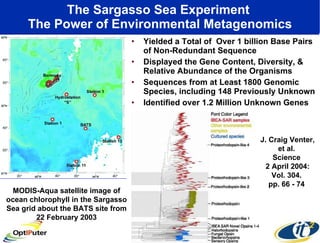
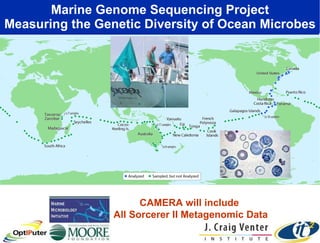
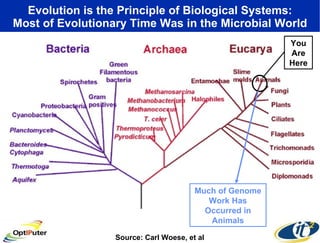
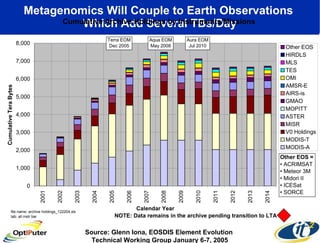
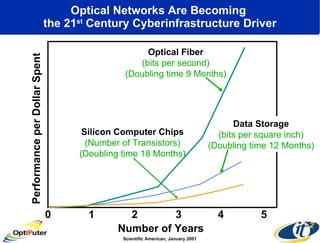
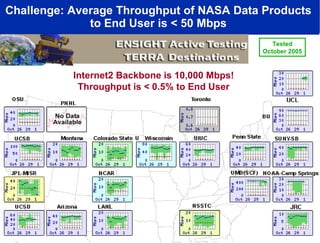
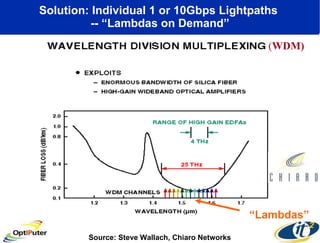
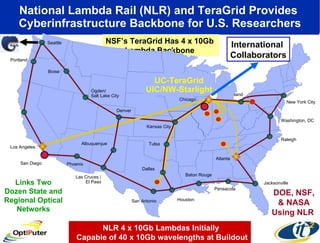
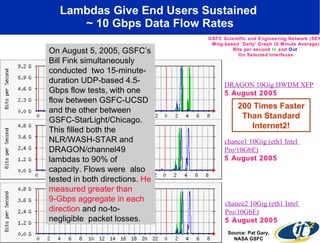
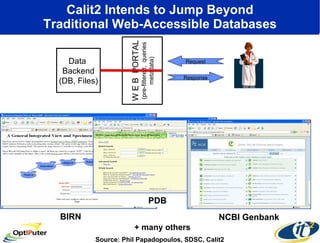
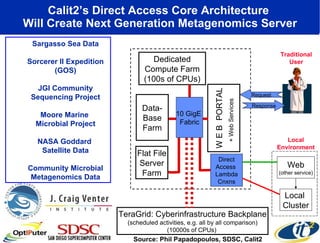
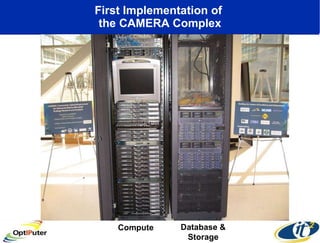
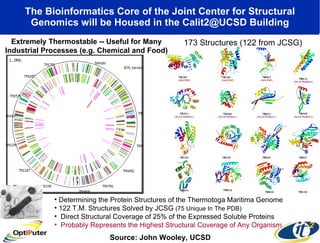
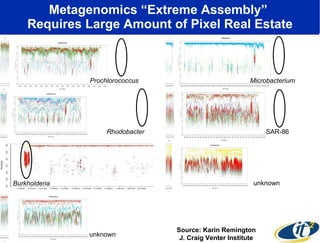
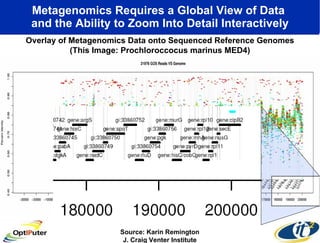
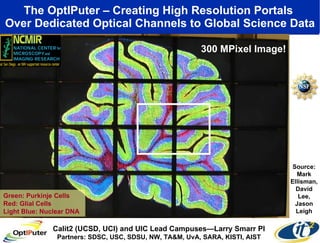
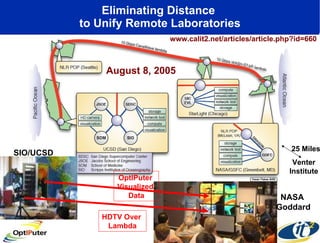
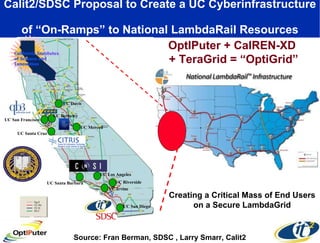
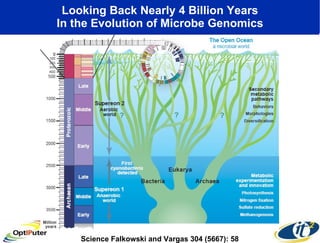
The document discusses advancements in microbial metagenomic sciences and the infrastructure necessary to support this research, particularly through the California Institute for Telecommunications and Information Technology. It highlights the Sargasso Sea experiment which has uncovered significant genetic diversity, and explains the technological advancements in data processing and transfer via optical networks, with a focus on the Optiputer project aimed at facilitating high-speed data analysis for large-scale genomic data. The presentation emphasizes the collaborative nature of the research community and the evolving understanding of microbial evolution.
![“ Building an Information Infrastructure to Support Microbial Metagenomic Sciences " Presentation for the Microbe Project Interagency Team [ www.microbeproject.gov] UCSD La Jolla, CA January 14, 2006 Dr. Larry Smarr Director, California Institute for Telecommunications and Information Technology; Harry E. Gruber Professor, Dept. of Computer Science and Engineering Jacobs School of Engineering, UCSD](https://image.slidesharecdn.com/microbeprojectjan2006final-100106160444-phpapp01/75/Building-an-Information-Infrastructure-to-Support-Microbial-Metagenomic-Sciences-1-2048.jpg)