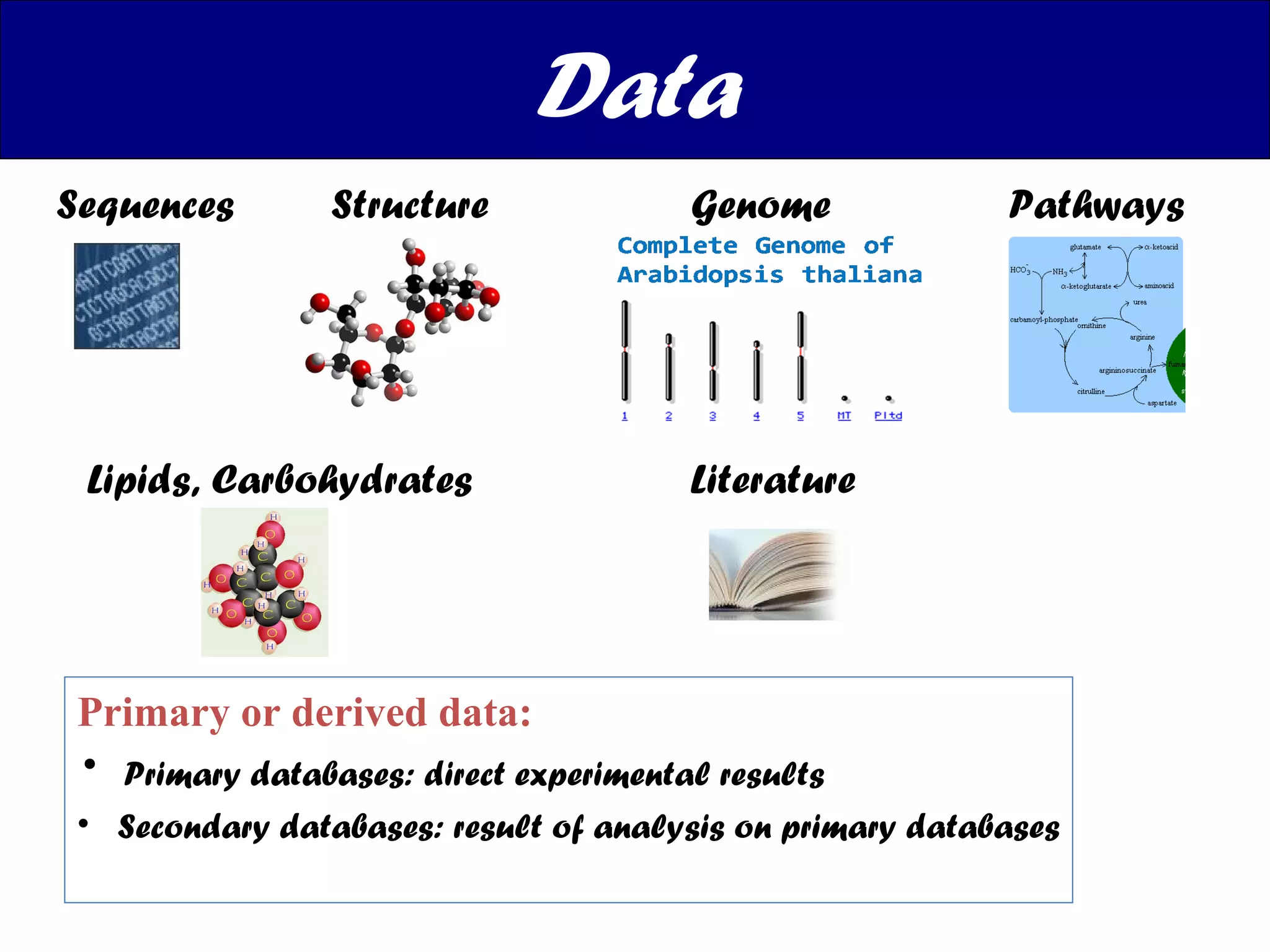
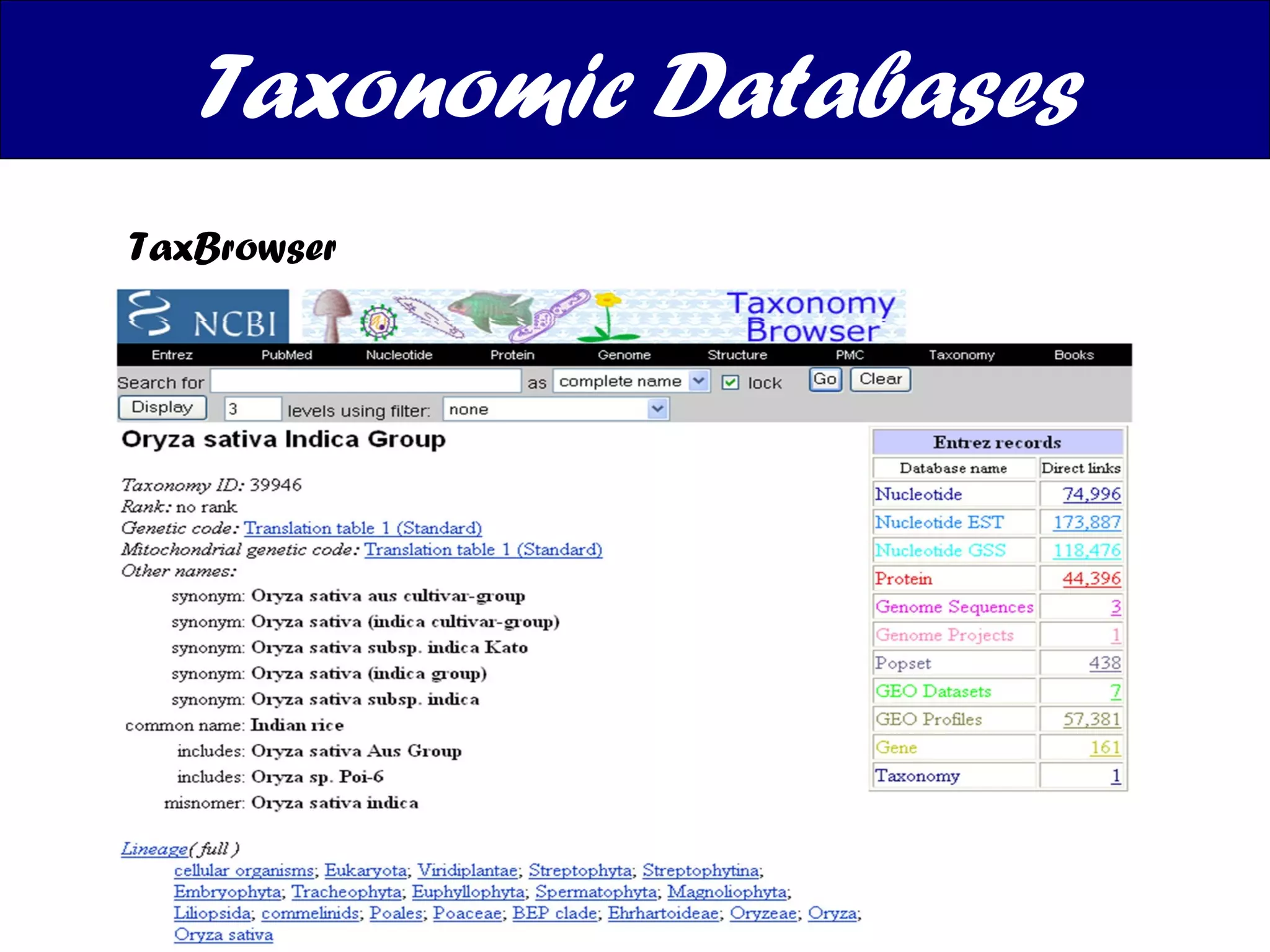
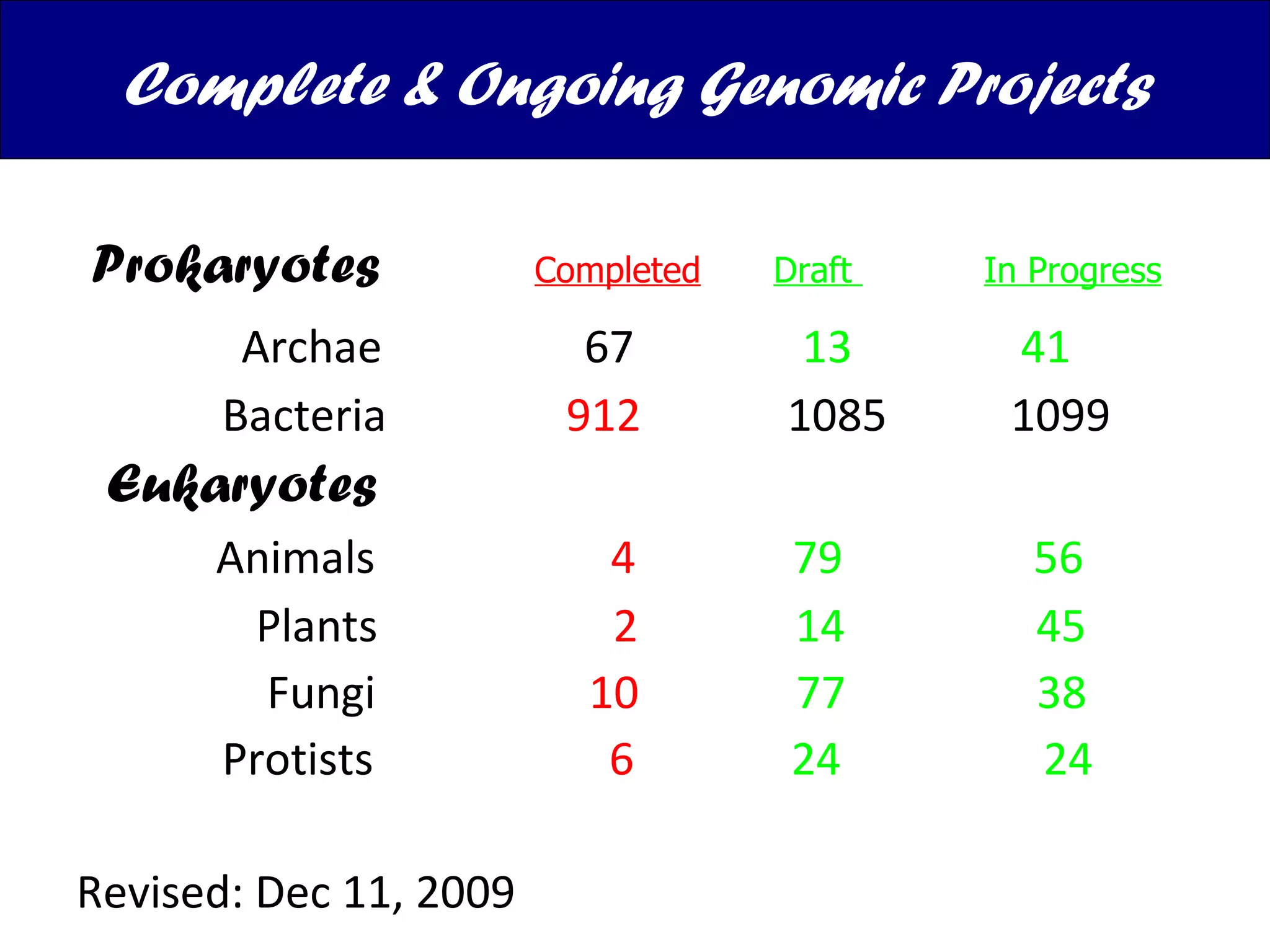
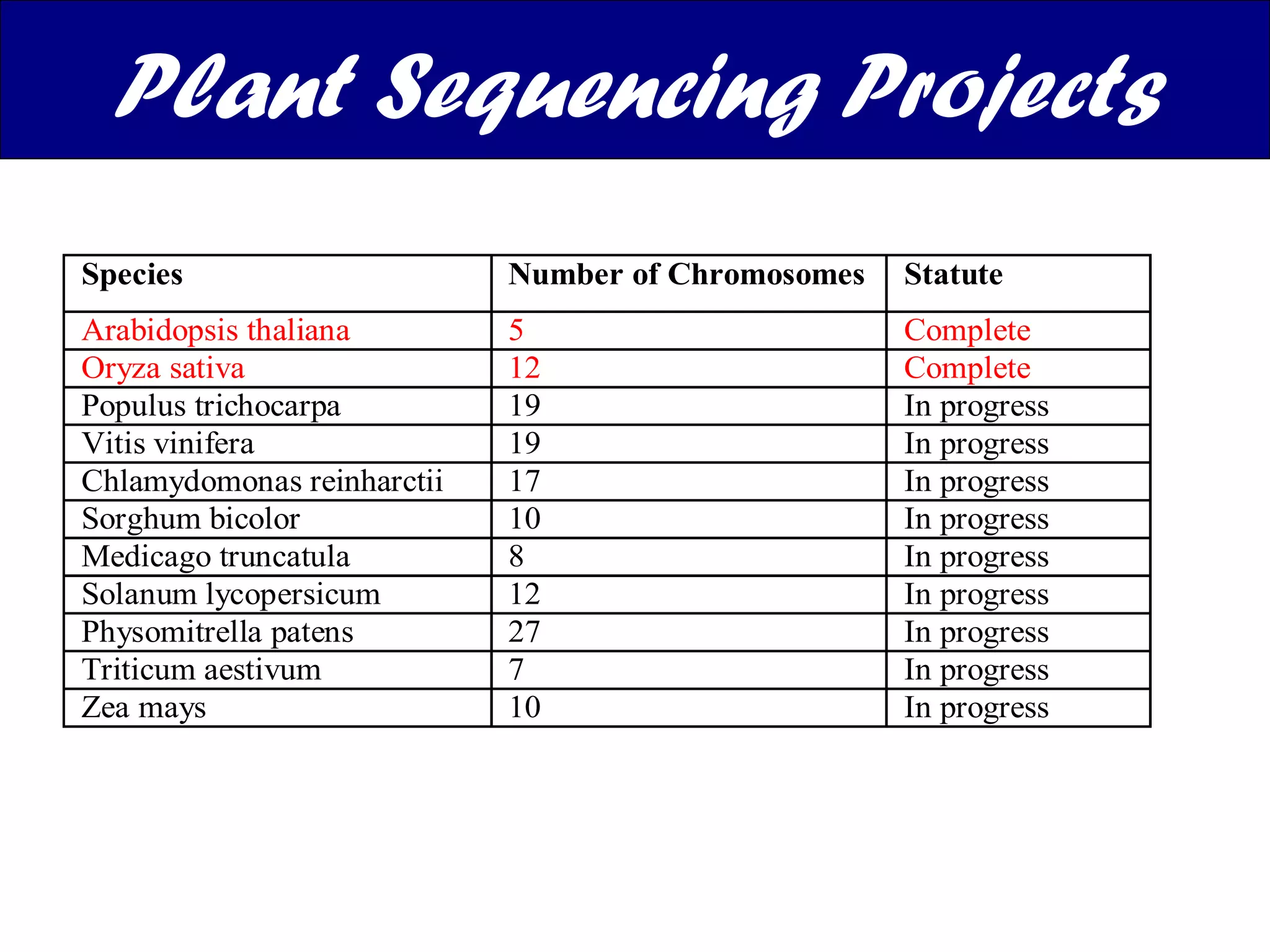
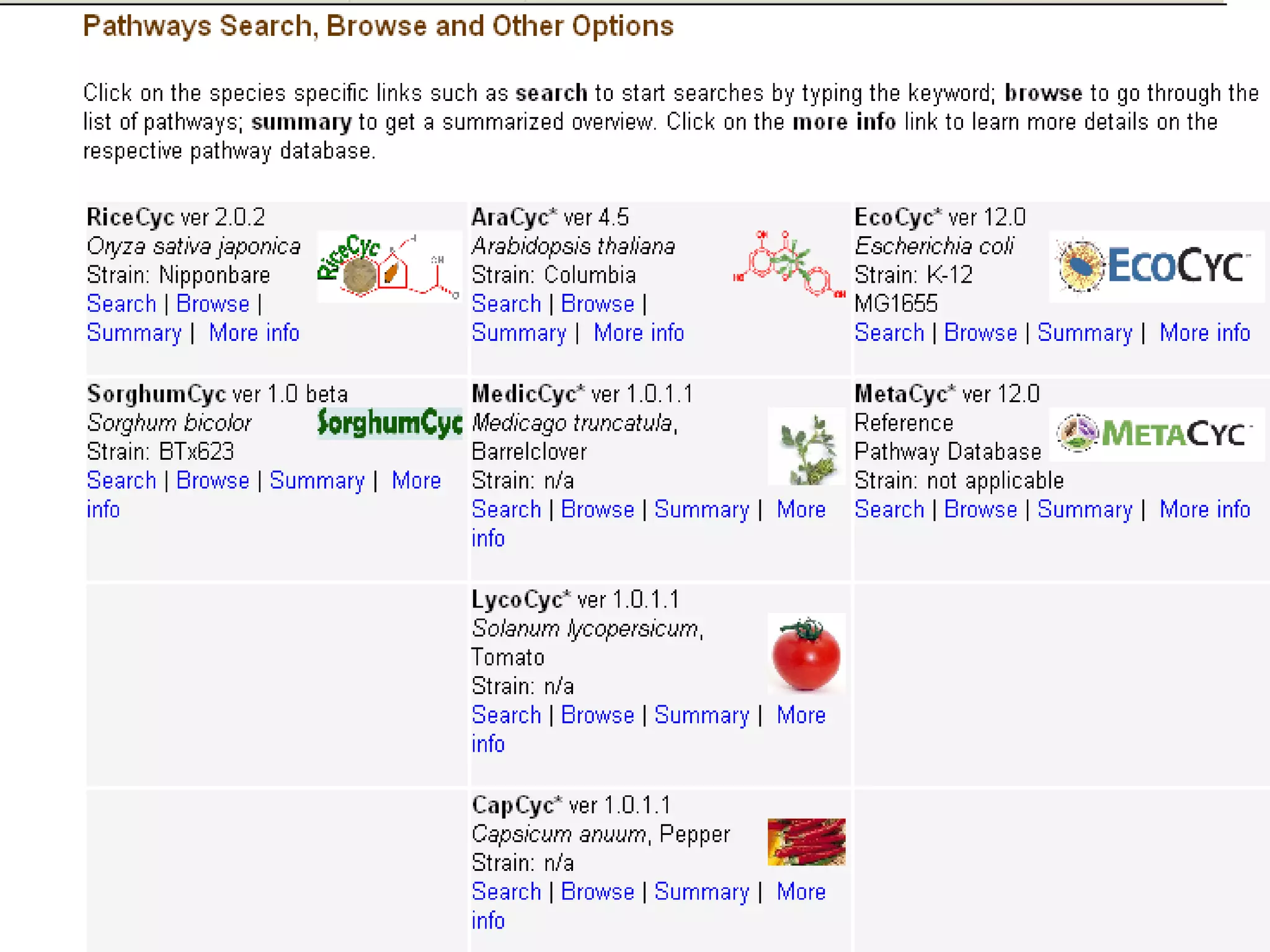
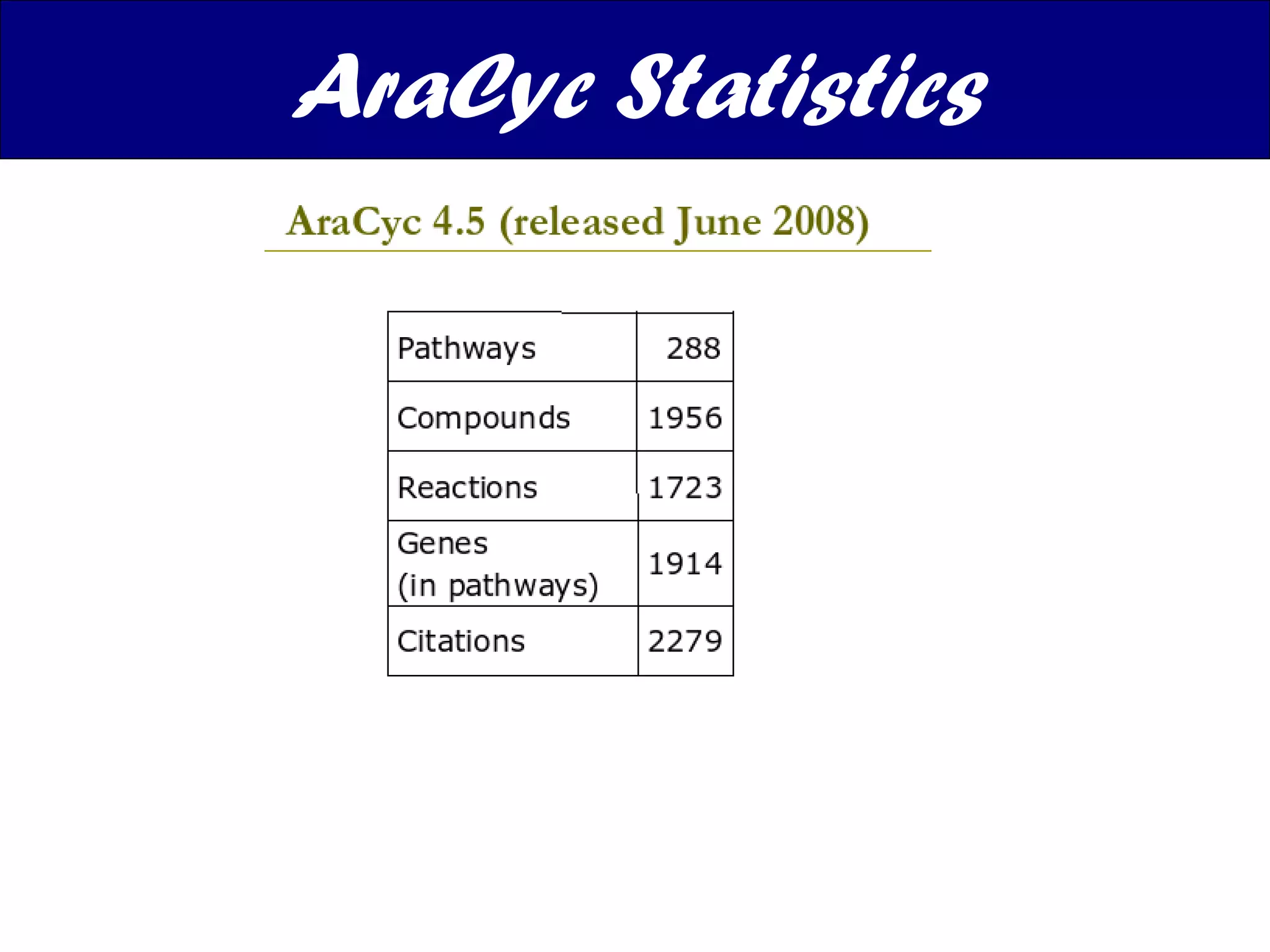
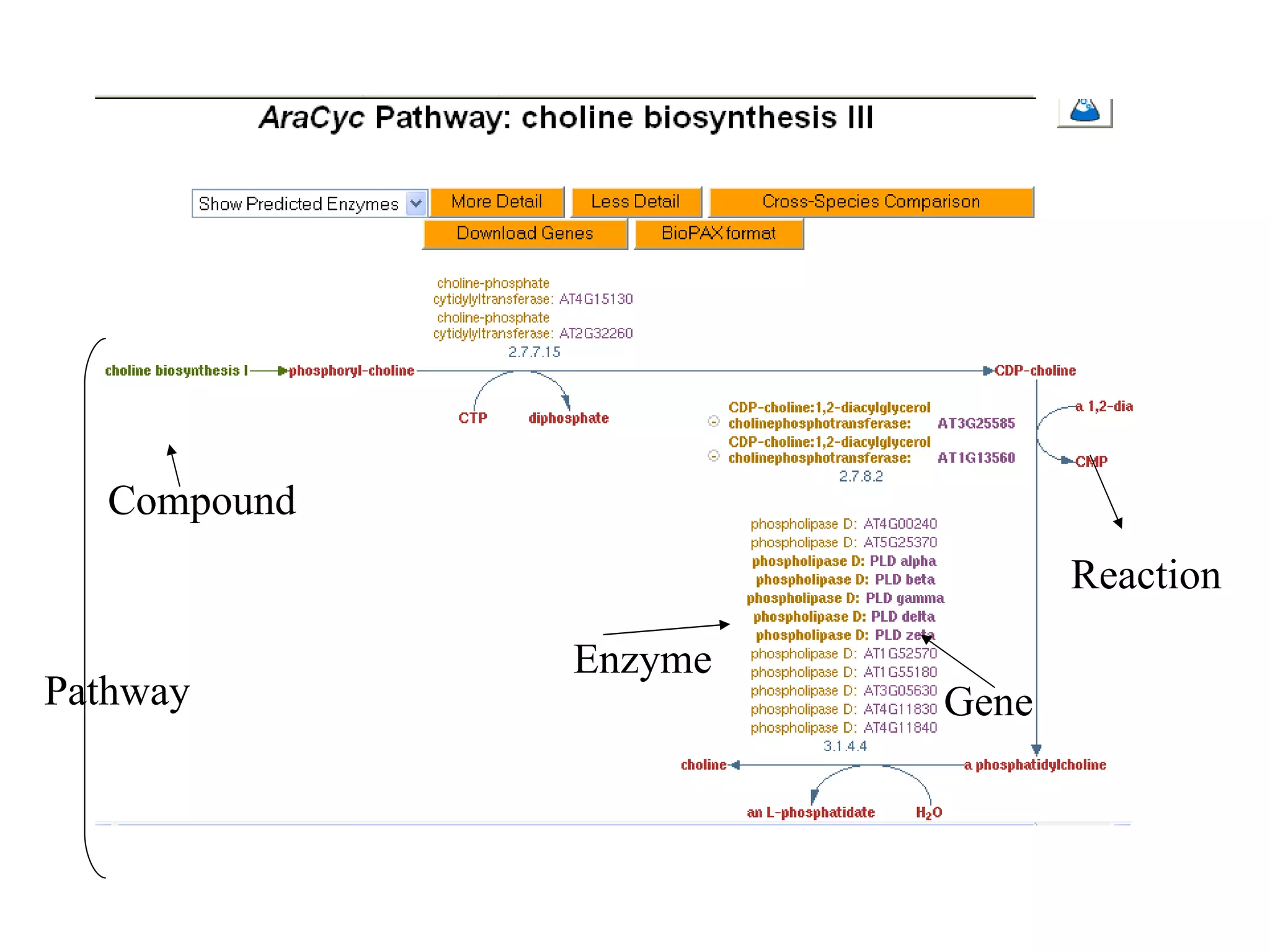
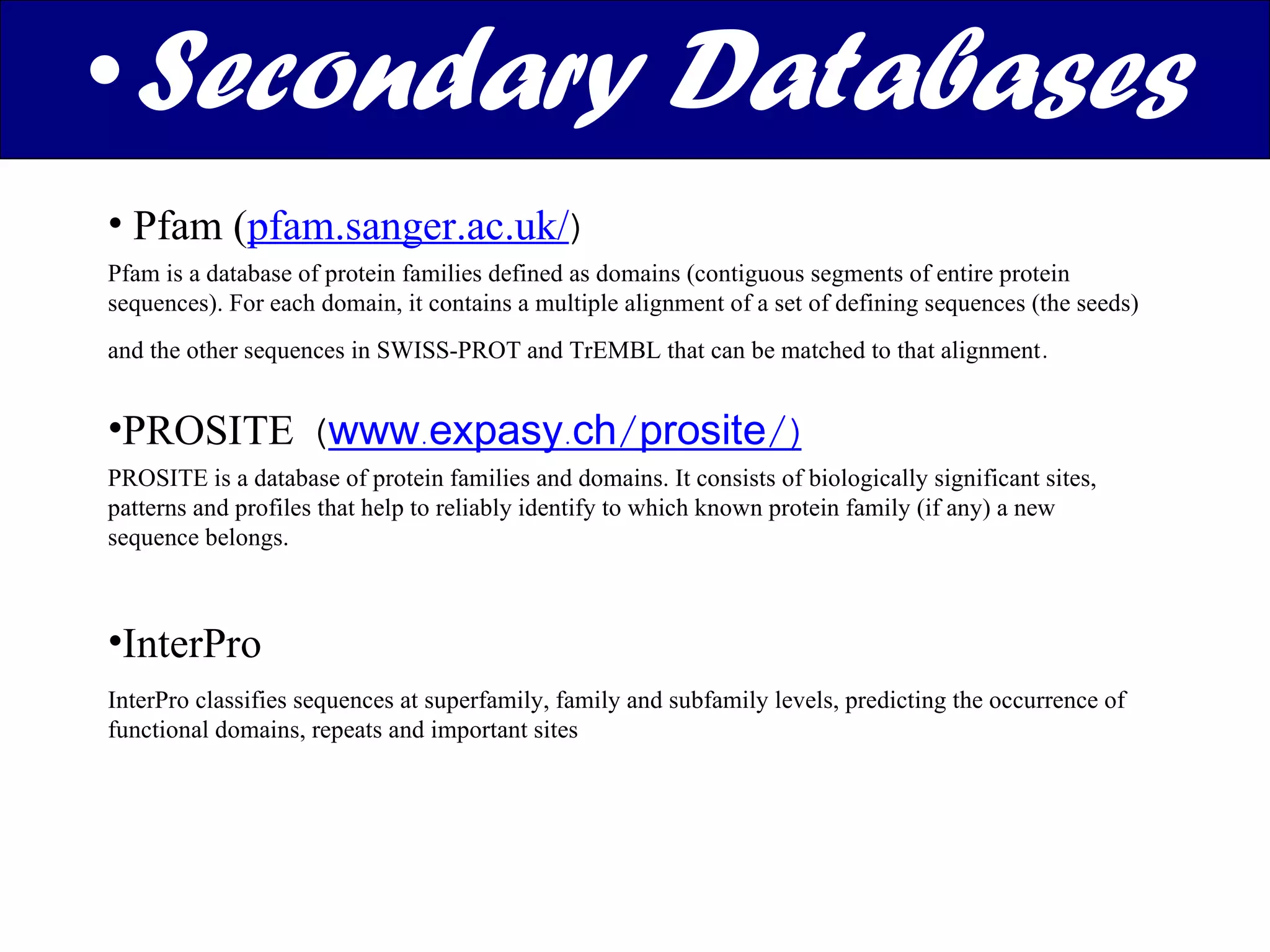
The document discusses various types of biological databases including nucleotide databases, genomic databases, protein databases, and metabolic databases. It provides examples of several specific databases, such as Nucleotide databases like GenBank, genomic databases like Entrez Genome, protein databases like UniProt, and metabolic databases like KEGG. It also discusses the different levels of data in biological databases from primary data directly from experiments to secondary data that is analyzed and derived from primary data.