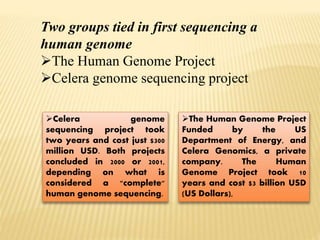
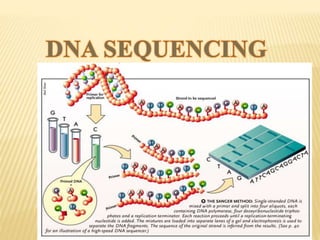
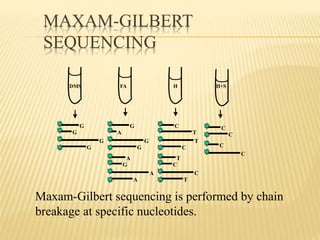
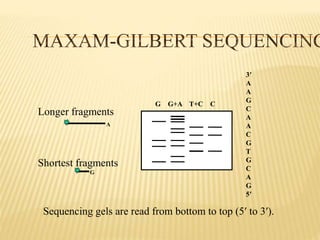
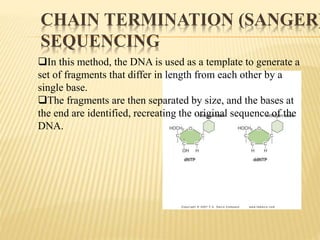
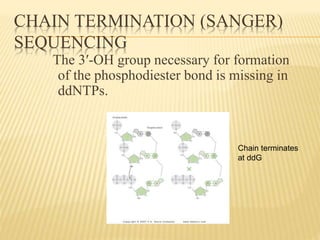
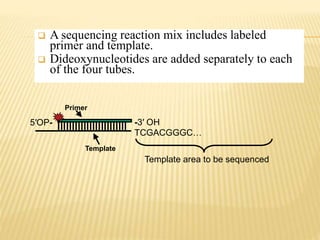
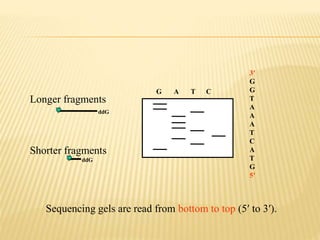
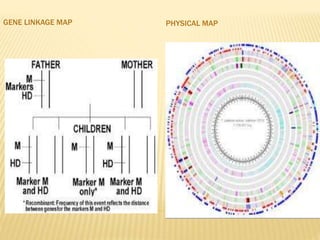
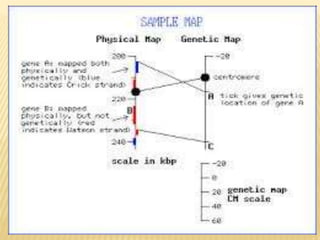
The Human Genome Project began in 1990 as a joint effort between the Department of Energy and National Institutes of Health to map the entire human genome. By 2001, both the publicly funded Human Genome Project and private company Celera Genomics had completed mapping the genome, which contains approximately 26,000 genes. DNA sequencing techniques like Sanger sequencing were used to determine the precise order of nucleotides in DNA and advance biological and medical research. The genome provides instructions to build a human through genes and proteins, with implications for medicine, biotechnology, and life sciences.