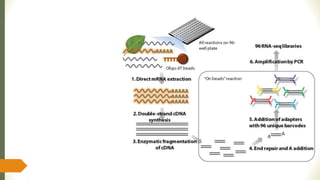
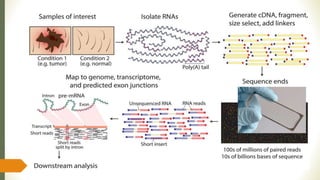
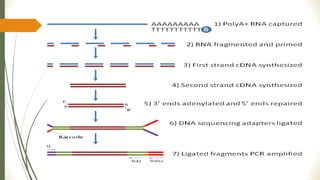
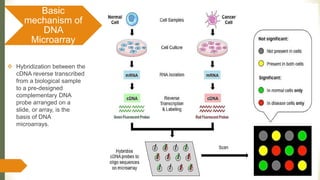
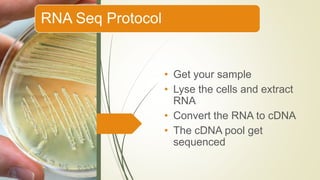
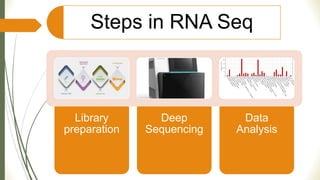
Gene expression and transcript profiling involves determining the pattern of genes expressed at the transcriptional level under specific circumstances by measuring the expression of thousands of genes simultaneously. This allows one to understand cellular function. Common techniques for profiling include DNA microarrays, RNA sequencing, and EST tags. DNA microarrays involve hybridizing cDNA or cRNA samples to probes on a chip to determine relative abundance of sequences. RNA sequencing uses next-generation sequencing to reveal presence and quantity of RNA in a sample.









![description smallmiRNA Degradome-
seq
Digital gene
expression
Poly[A]-RNA
Seq
Total RNA seq
Library
preparation-
RNA selection
Size selection-
small RNA
Poly[A] tail
selection-
miRNA
Poly[A] tail
selection-
mRNA
Poly[A] tail
selection-
mRNA
rRNA depletion
-
mRNA+IncRNA
Sequencing
cycles
50 50 50 2×100 2×100
Single vs.
paired end
Single end Single end Single end Paired end Paired end
Data analysis discovery
expression
annotation
miRNA target
ID degradation
plots
Expression
annotation
discovery
expression
annotation
discovery
expression
annotation](https://image.slidesharecdn.com/geneexpressionprofiling-181013111841/85/Gene-expression-profiling-13-320.jpg)