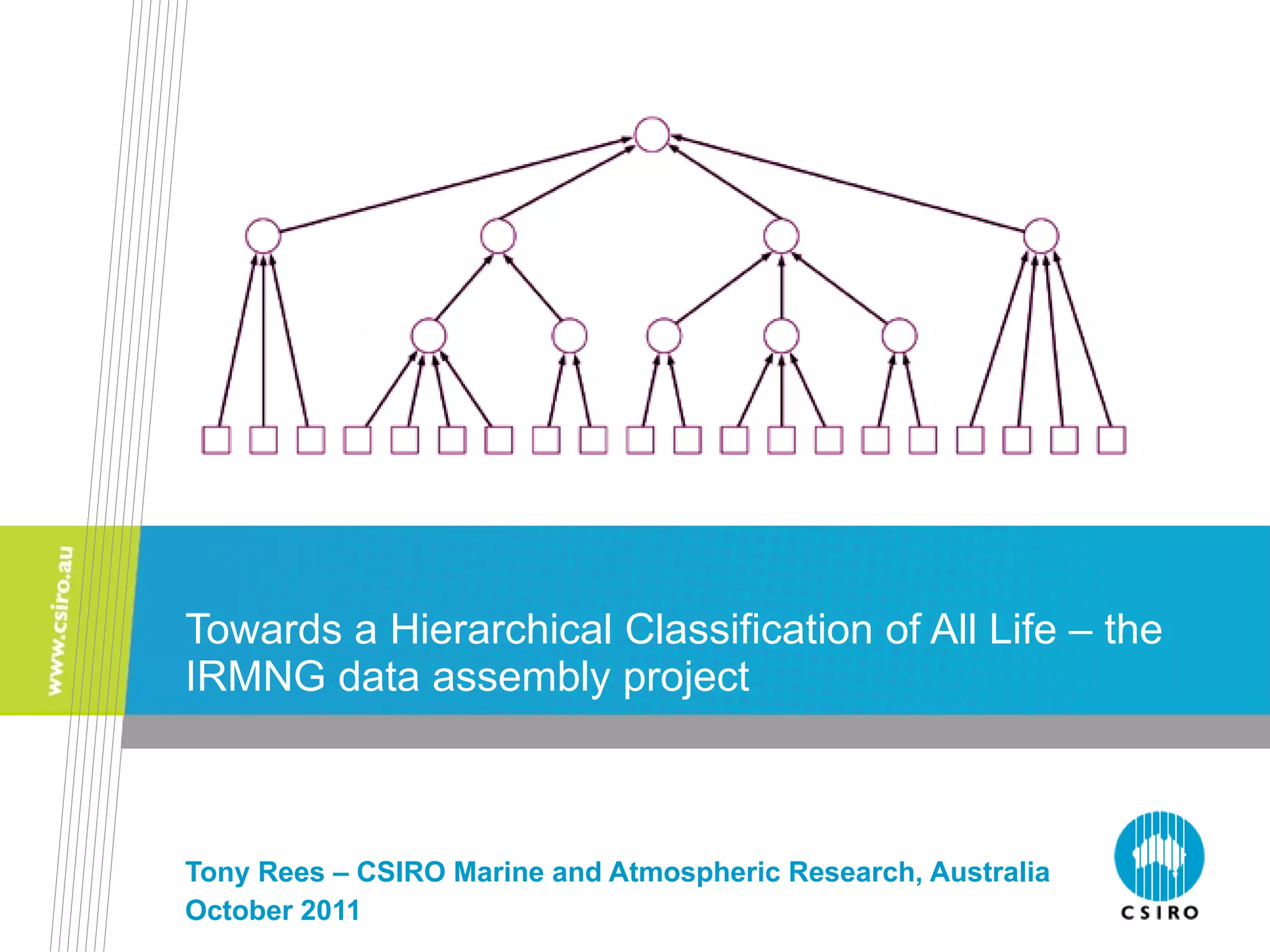
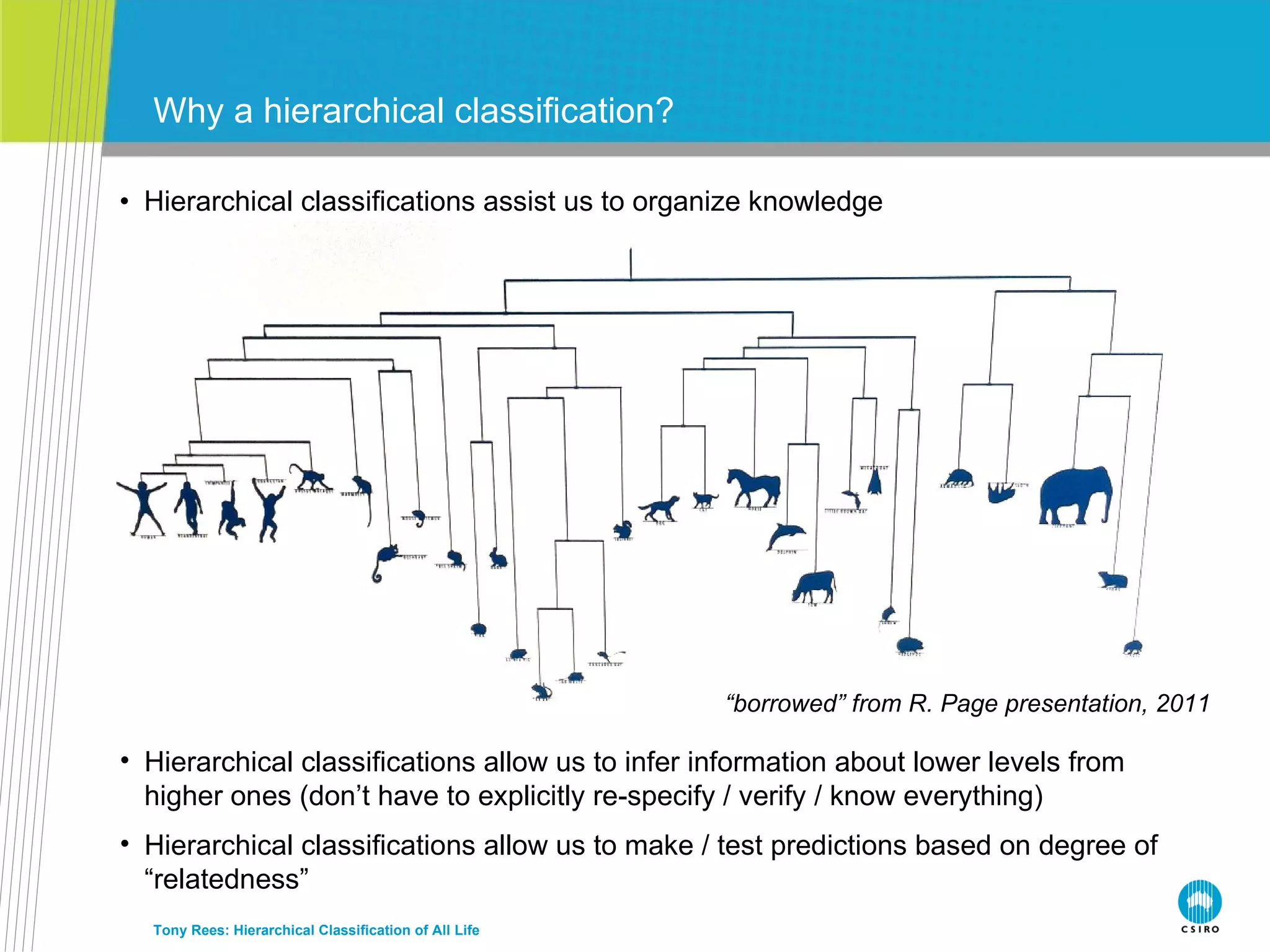
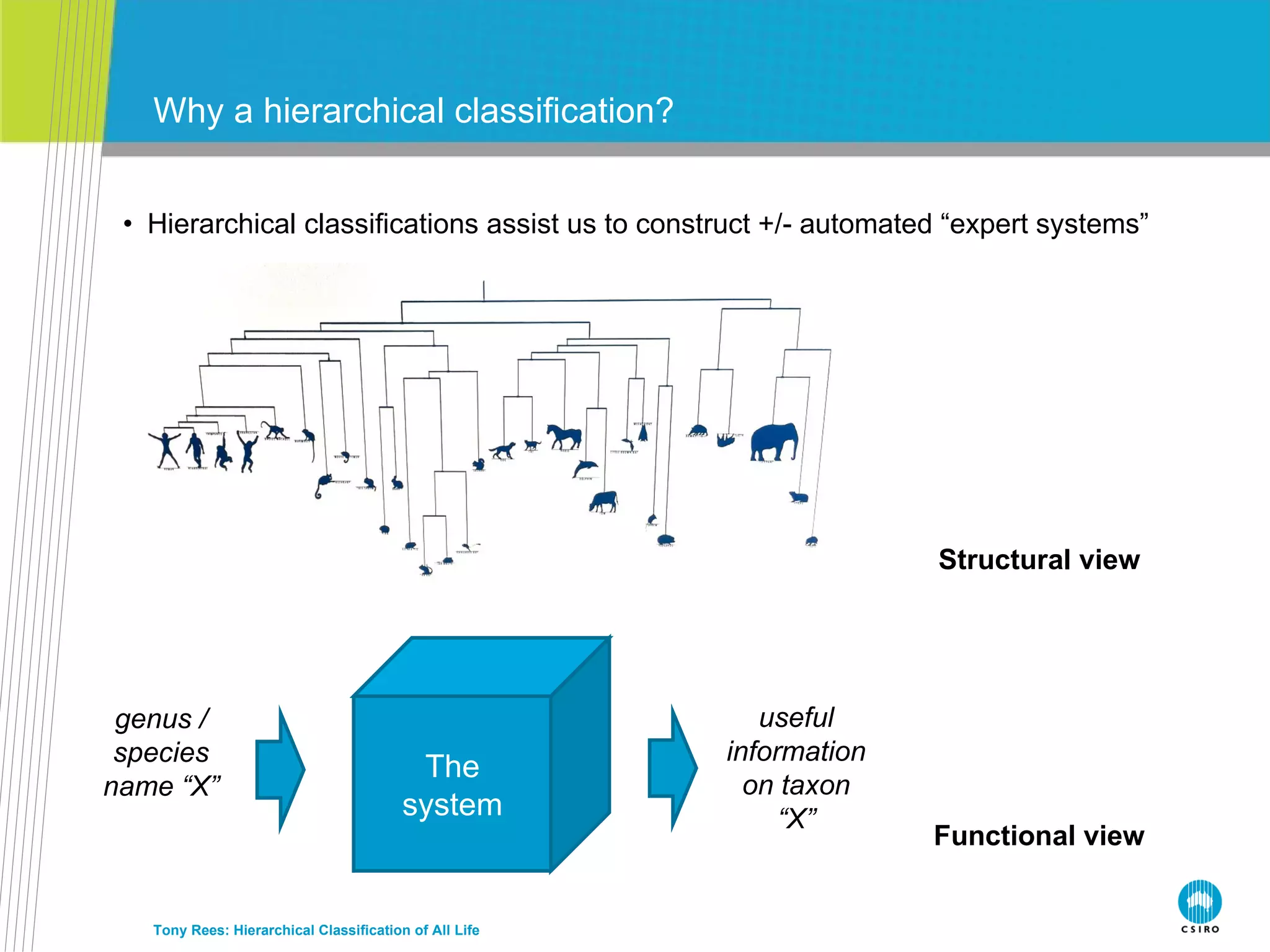
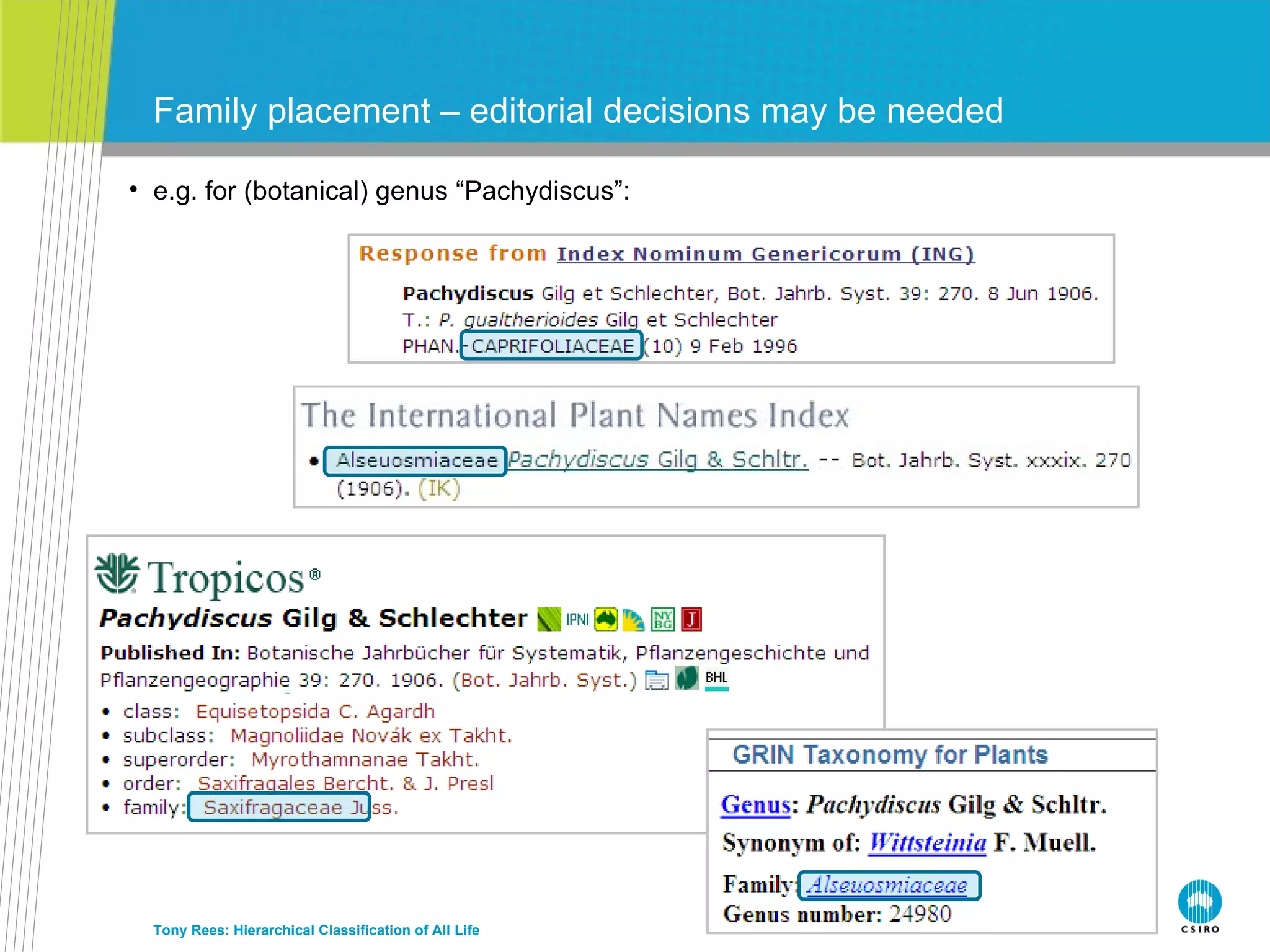
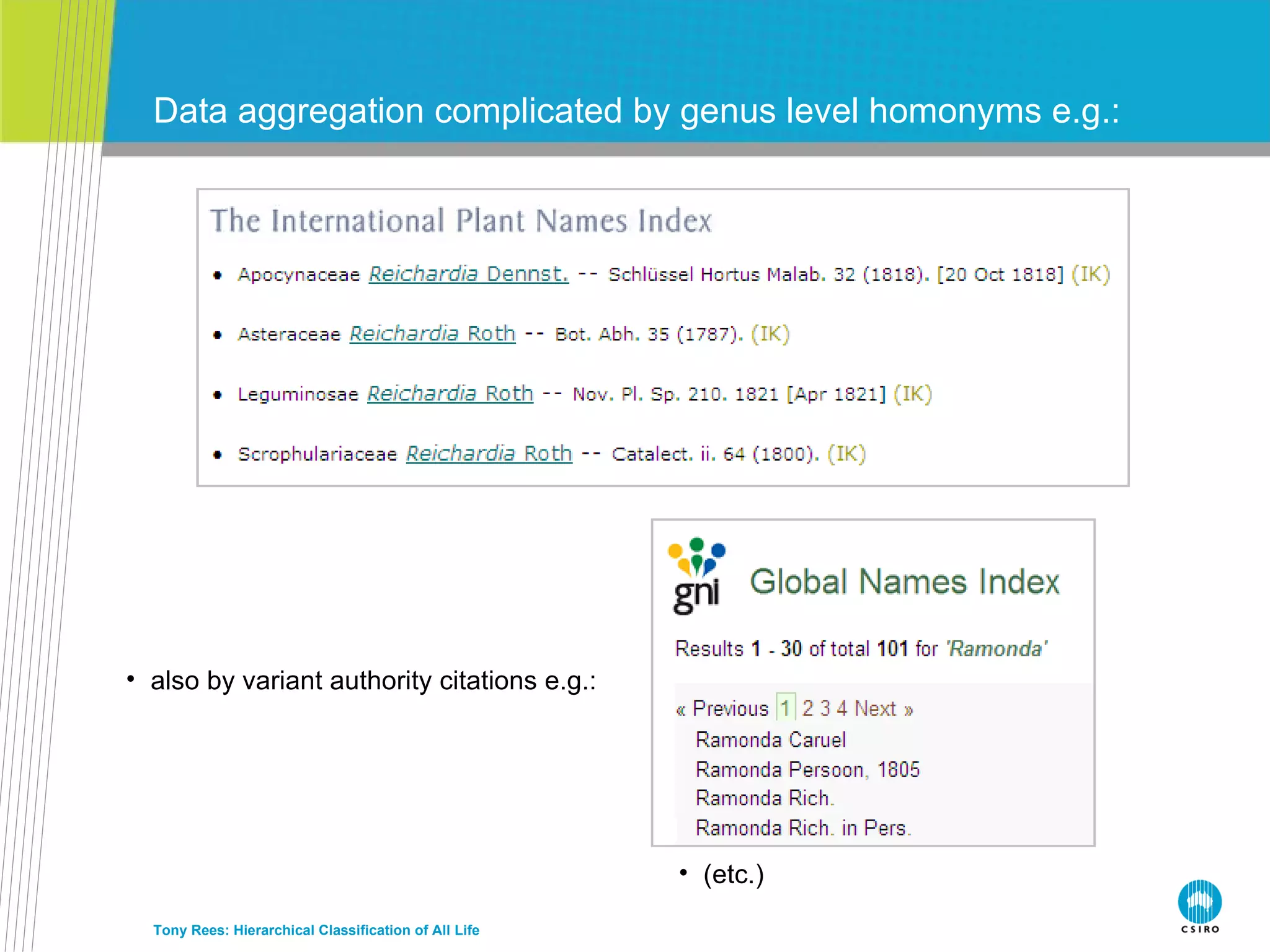
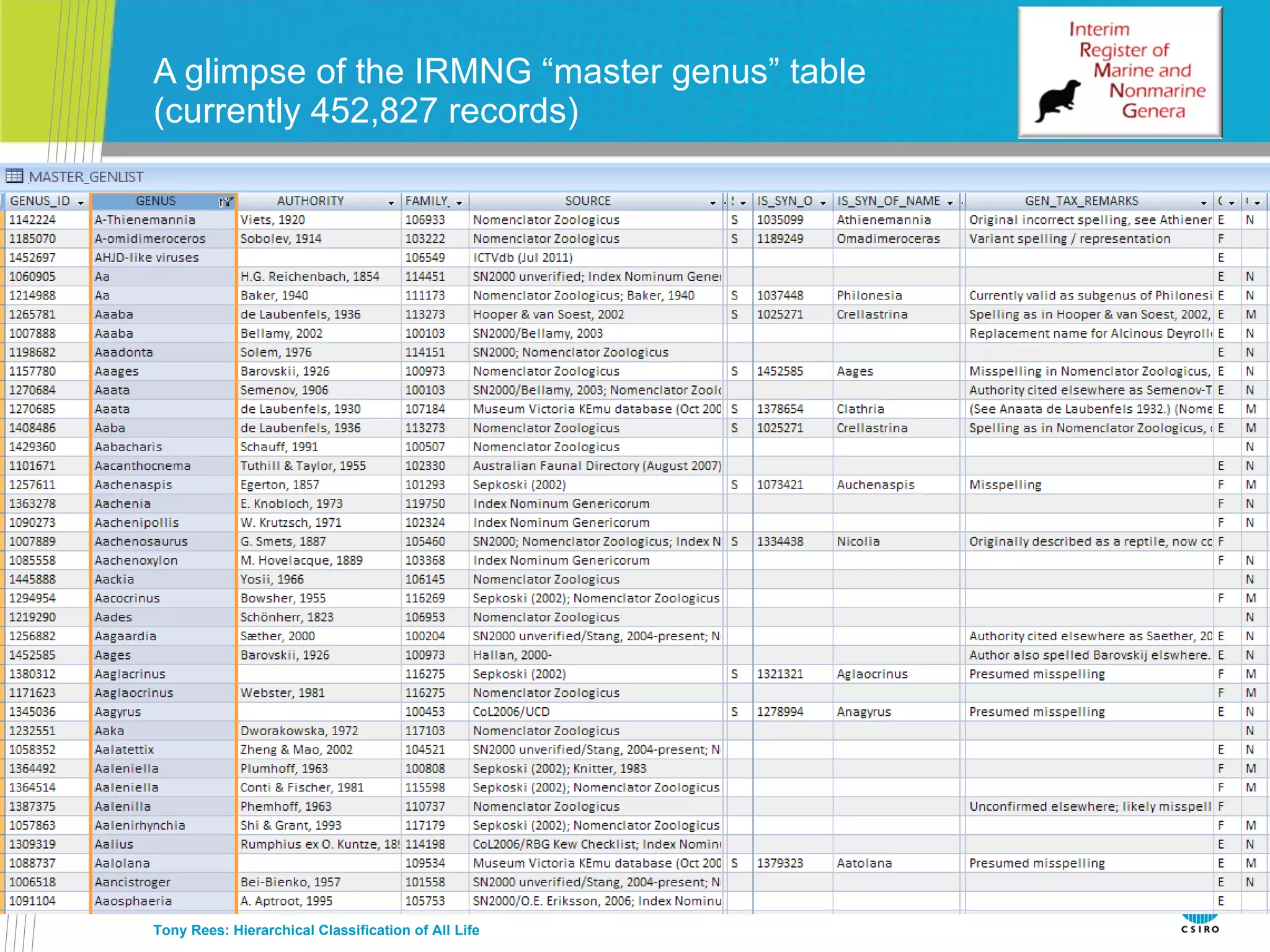
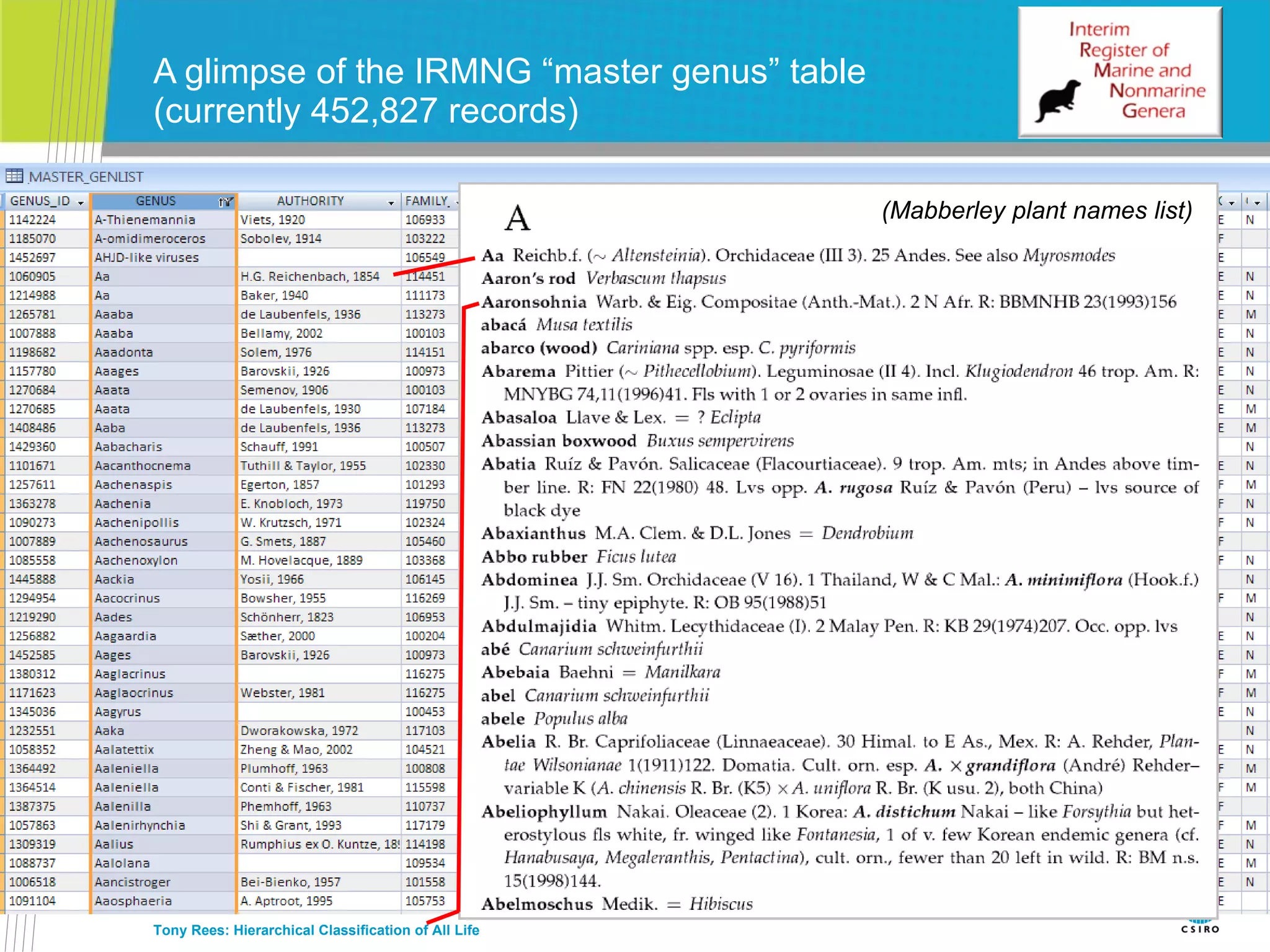
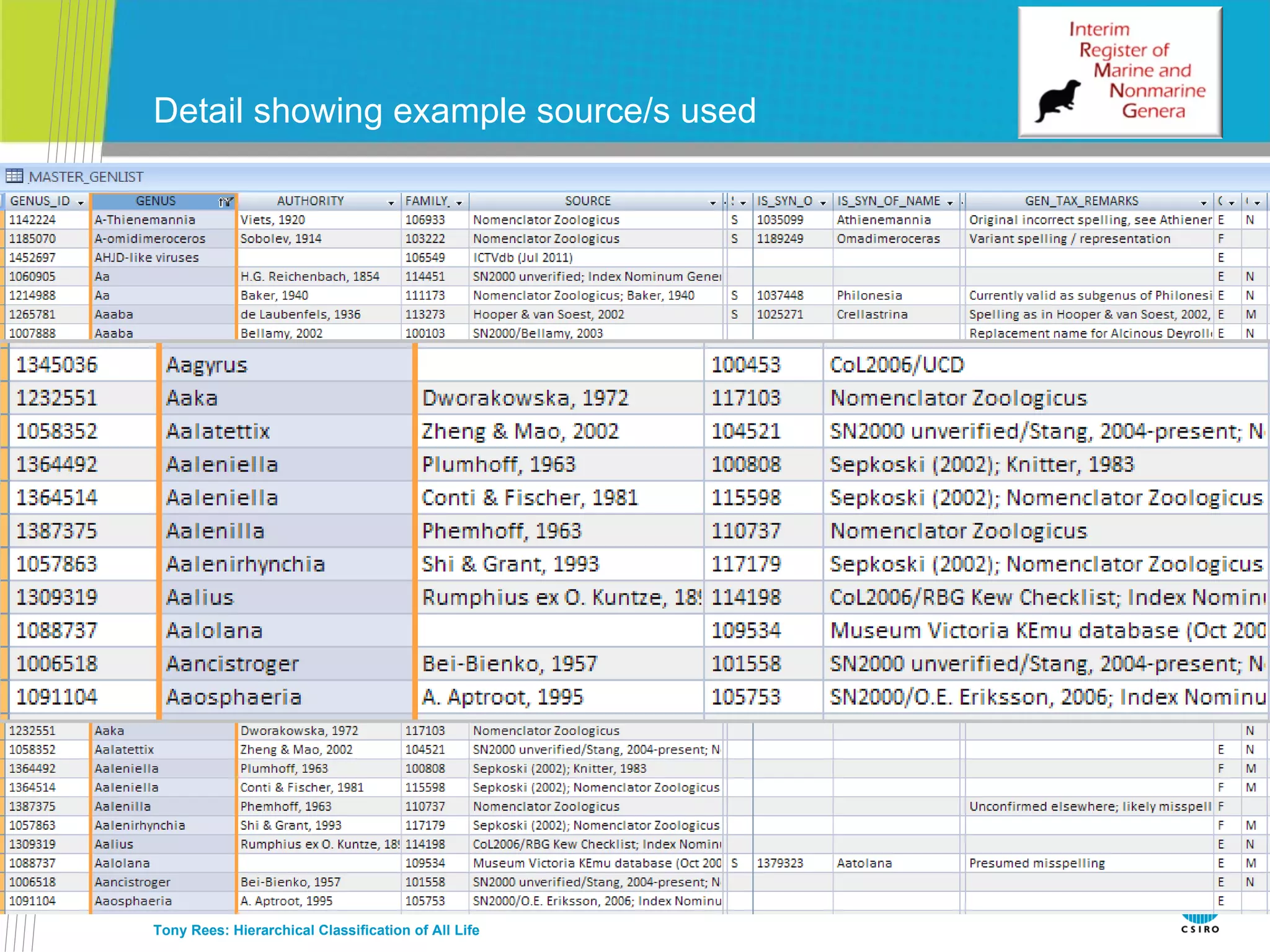
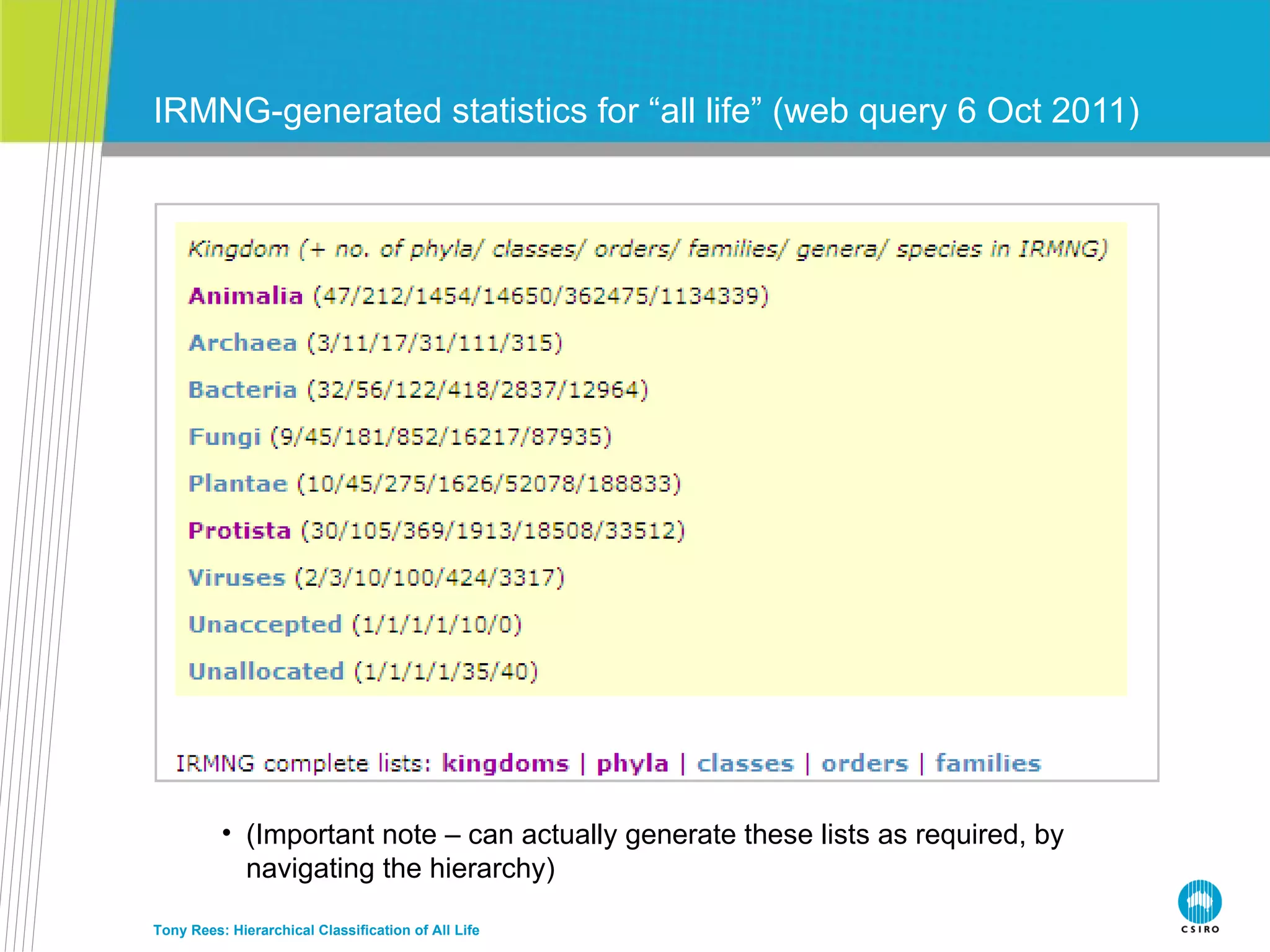
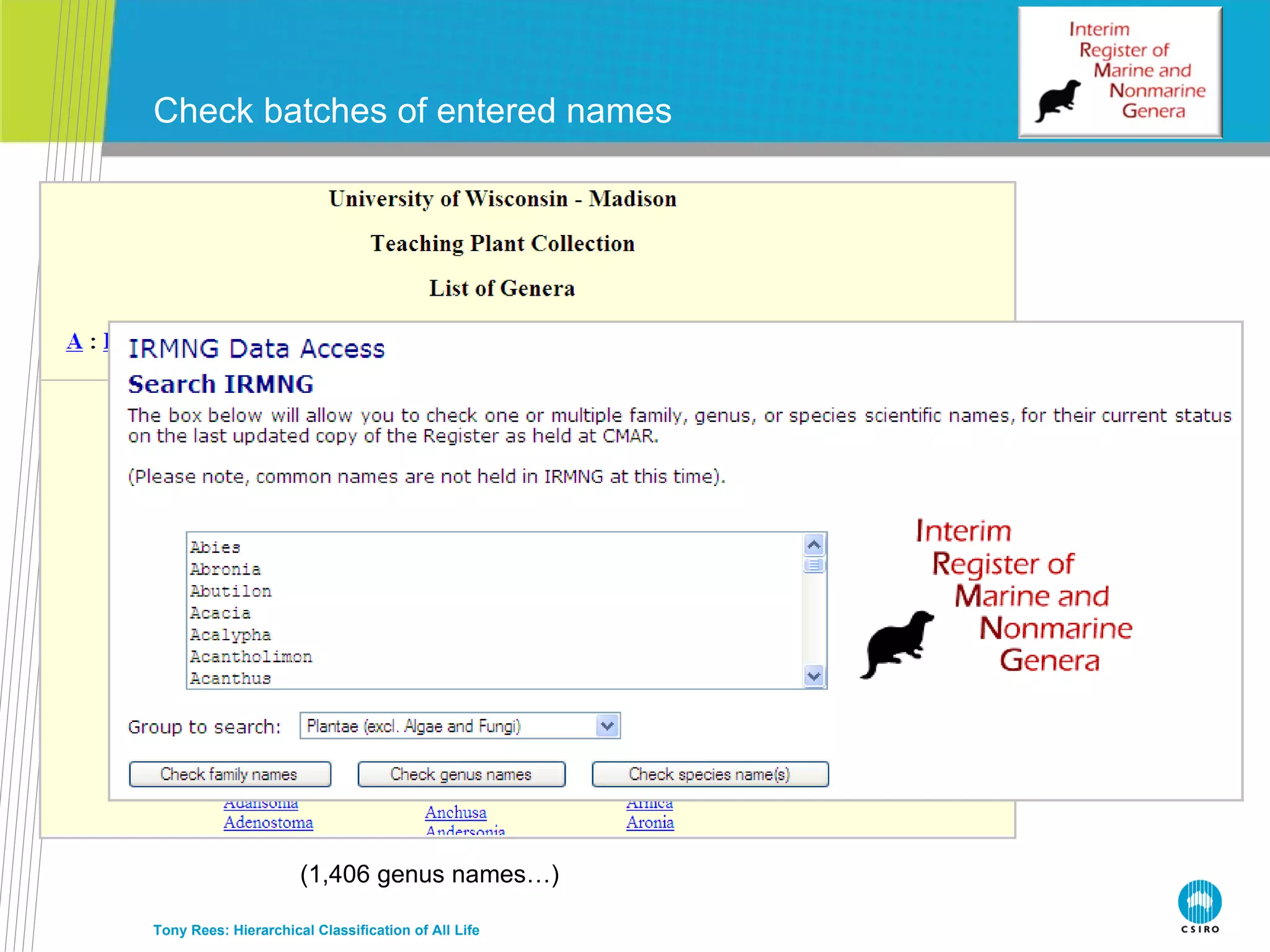
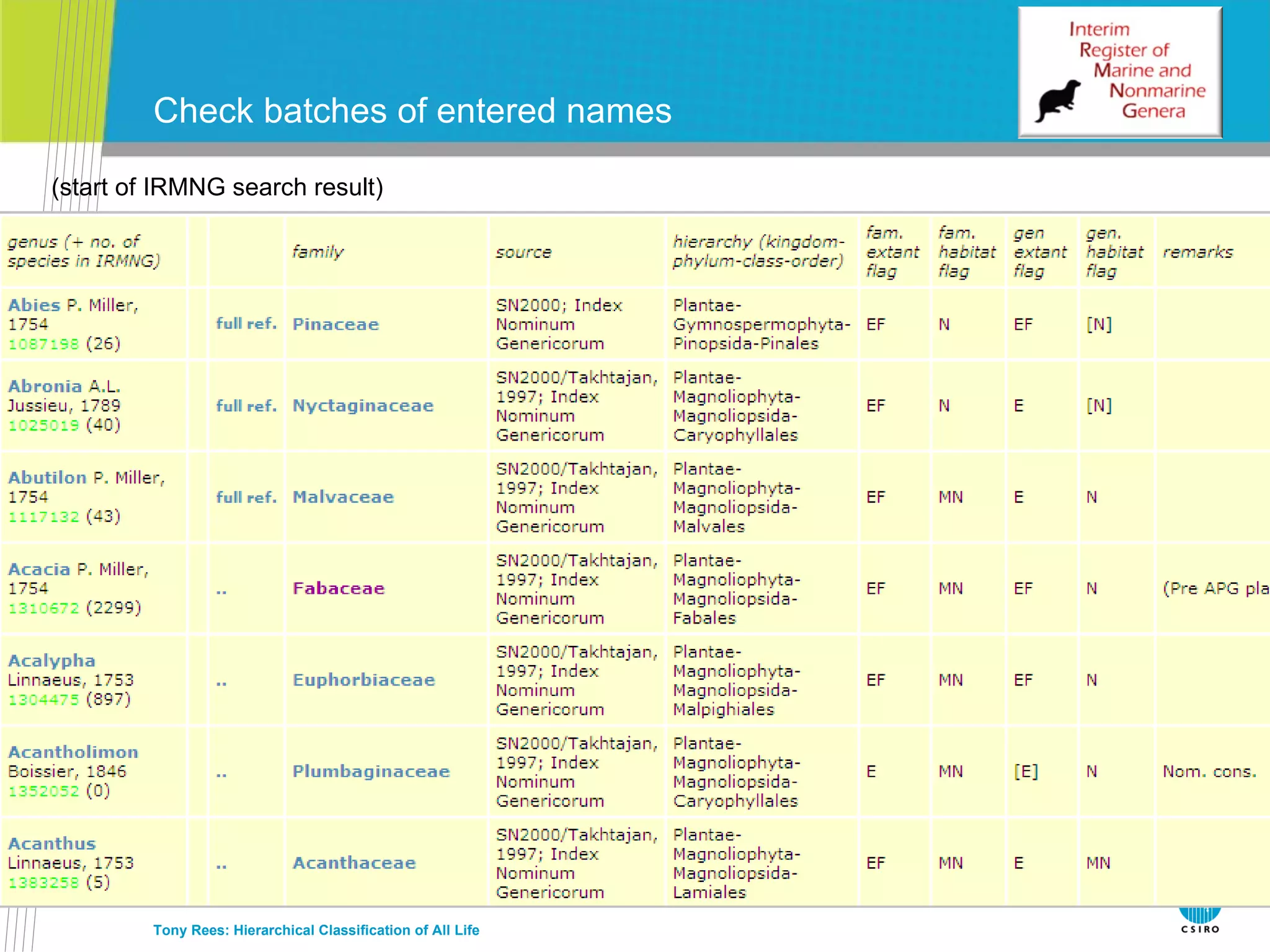
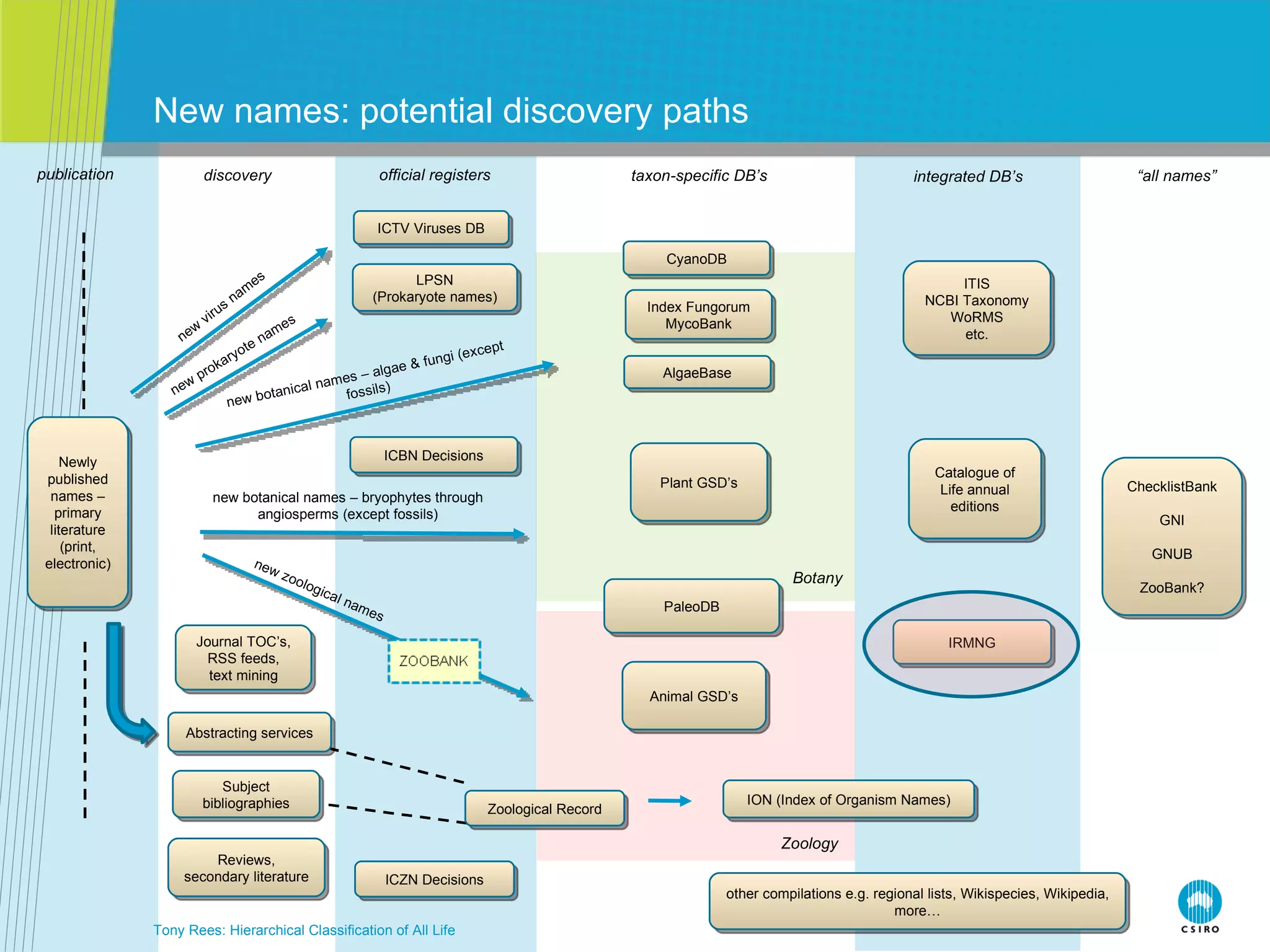
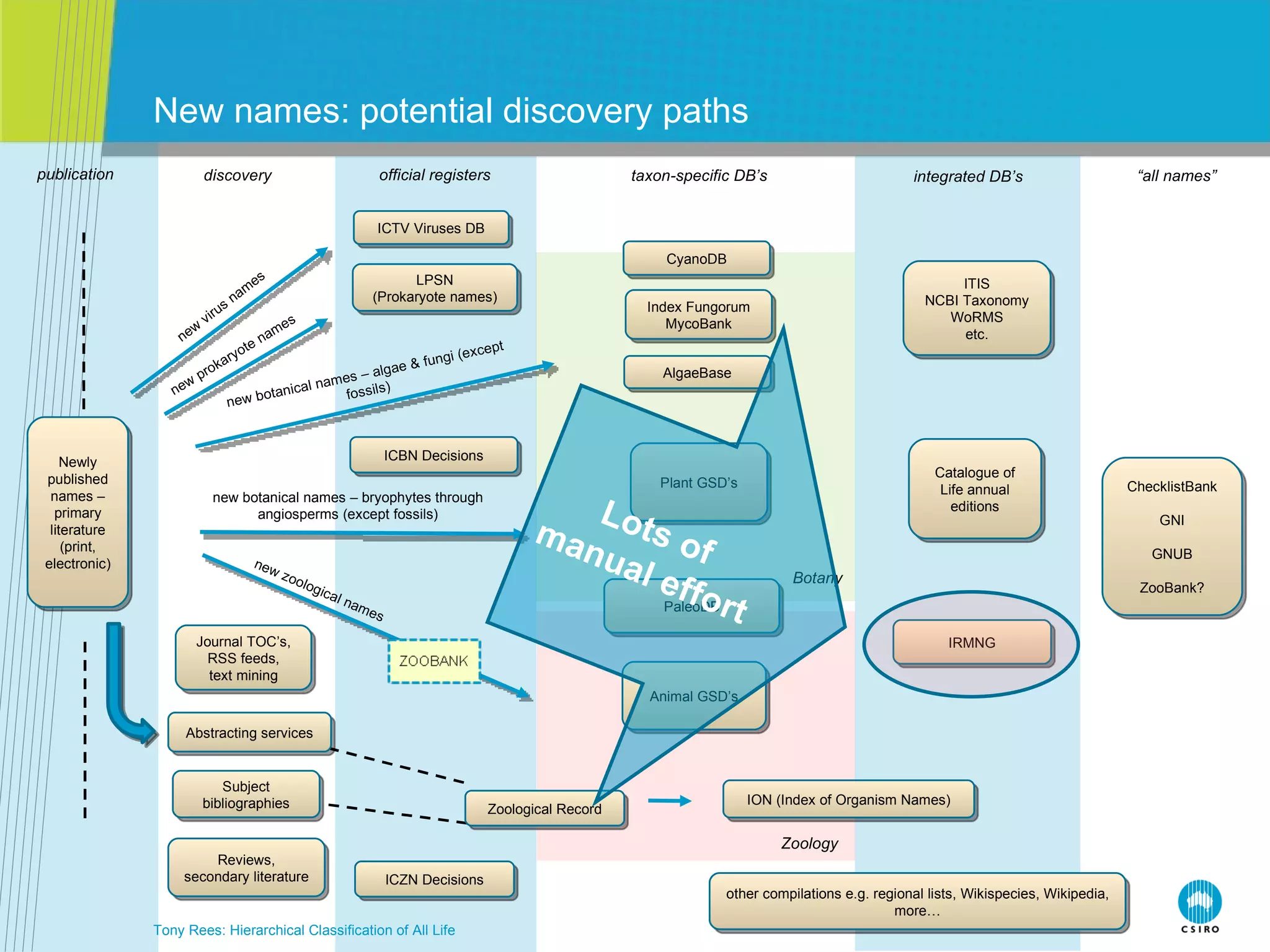
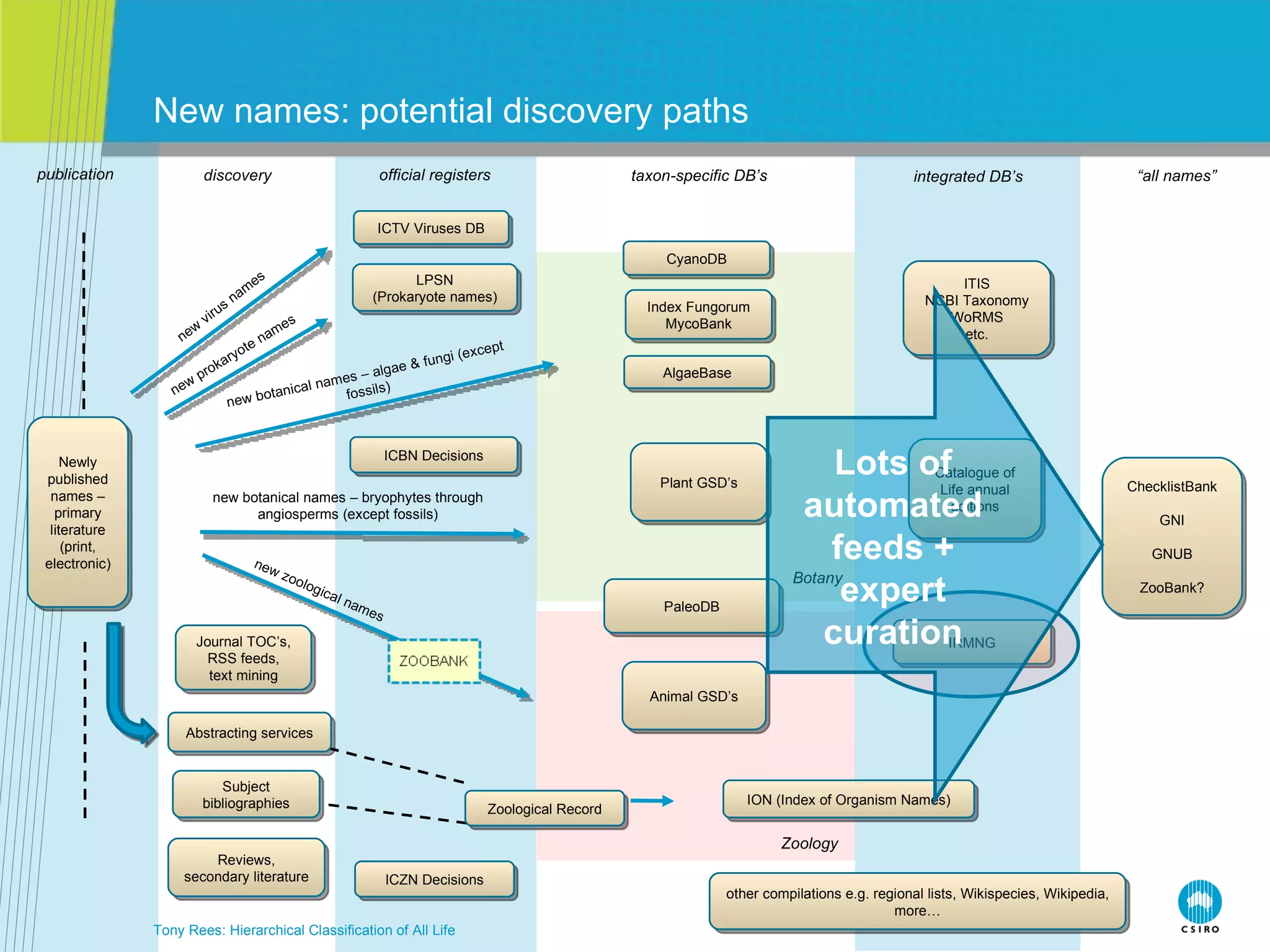
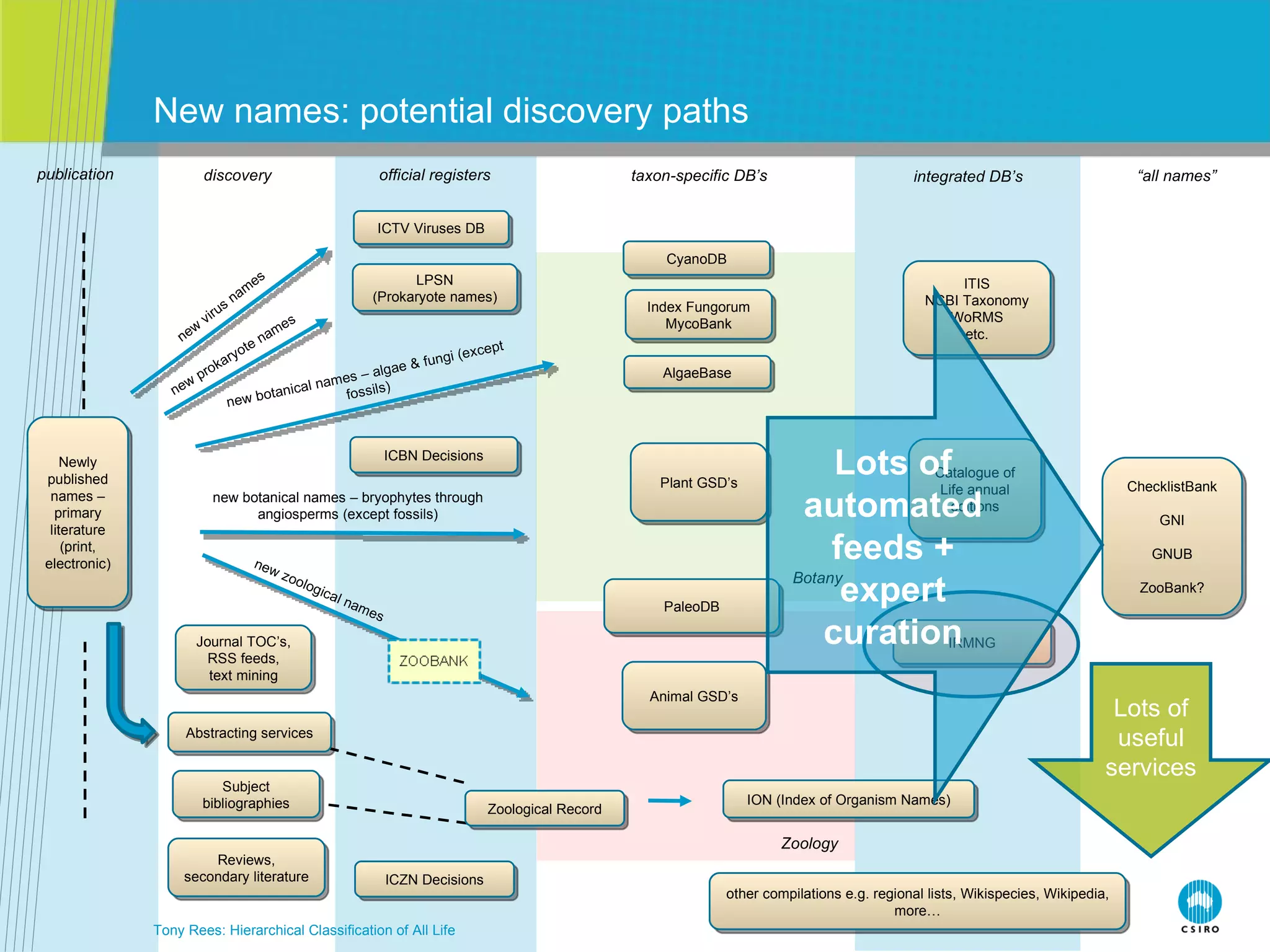
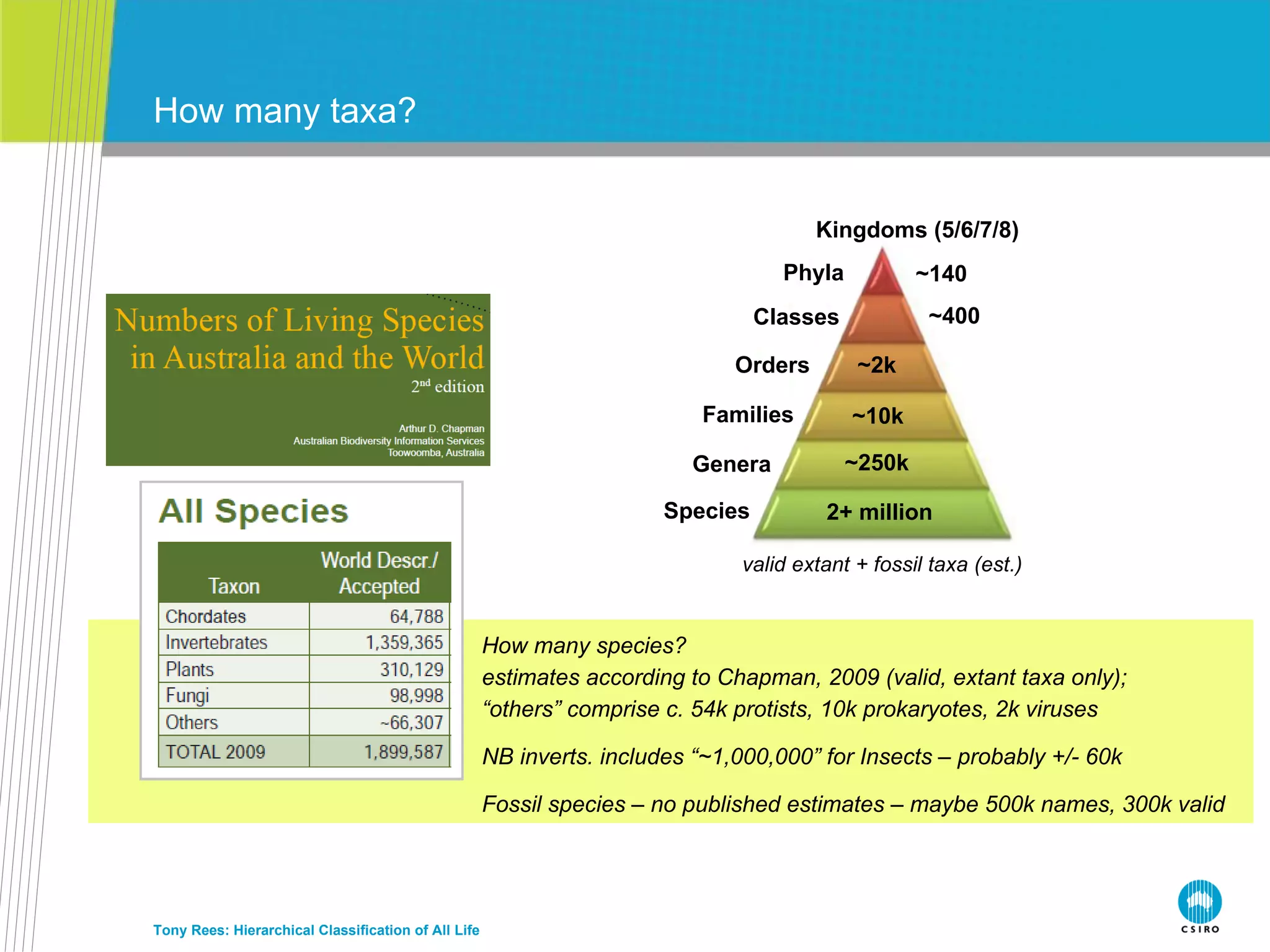
The document discusses the IRMNG (Interim Register of Marine and Nonmarine Genera) data assembly project, which aims to create a hierarchical classification of all life, both extant and fossil, to at least the genus level. It provides an overview of the goals and structure of IRMNG, which compiles name and taxonomic data from various sources into a single, searchable system. IRMNG currently contains over 450,000 genus records and aims to fill gaps, resolve inconsistencies between sources, and serve as a comprehensive yet interim reference for the names and classification of all organisms.
















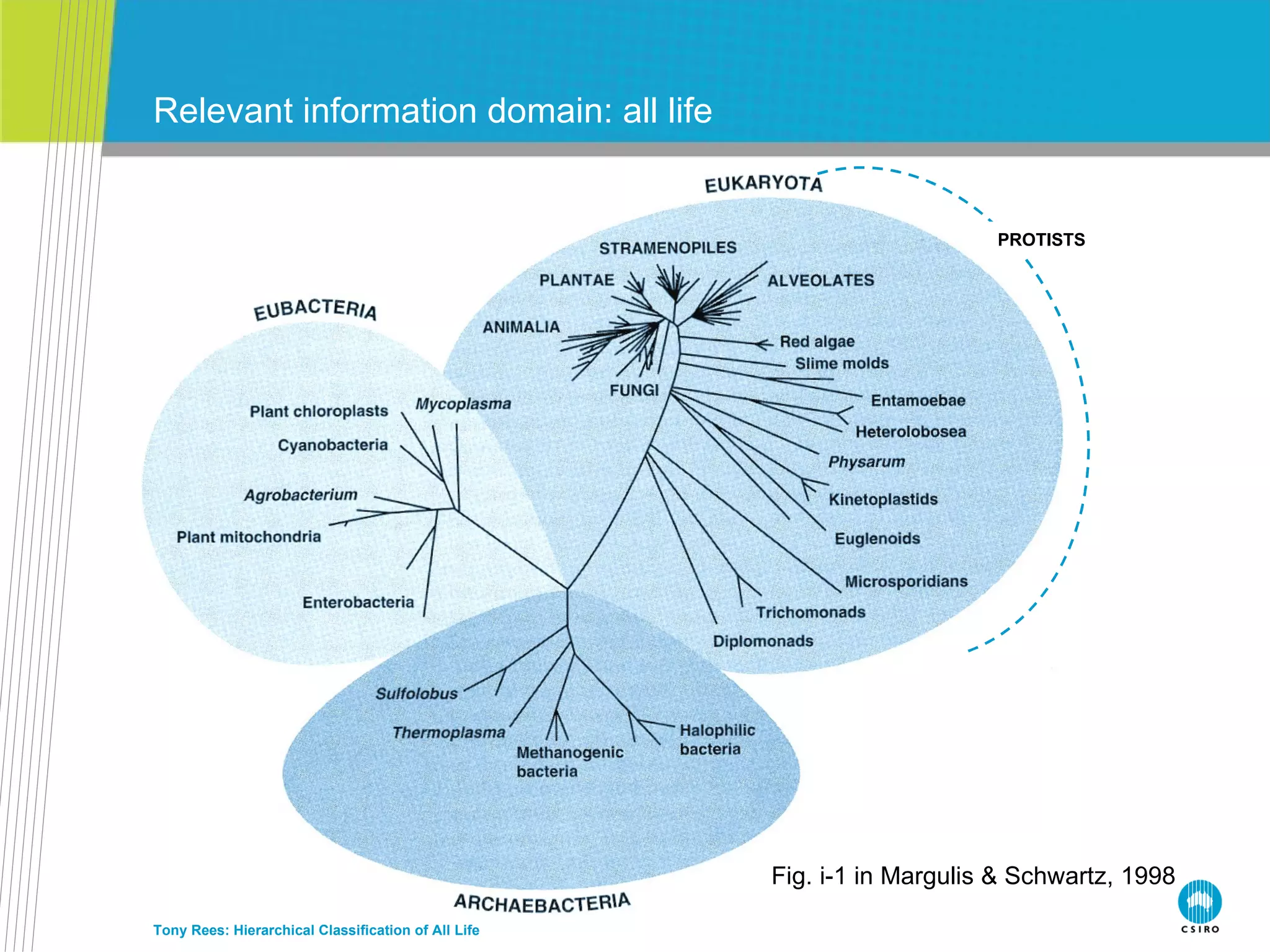
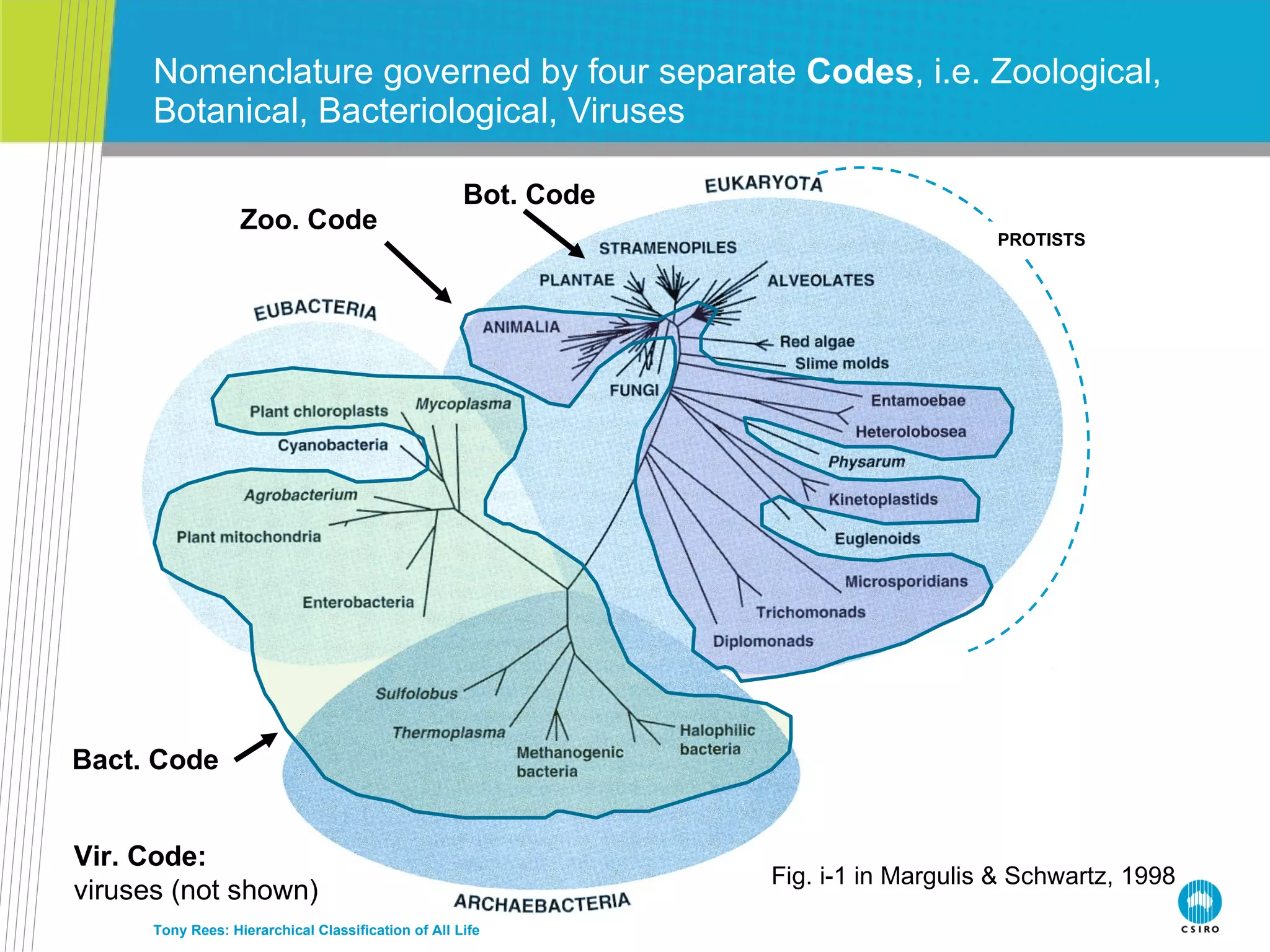
![How many kingdoms… Tony Rees: Hierarchical Classification of All Life PROTISTS Fig. i-1 in Margulis & Schwartz, 1998 7 kingdoms (5 in Margulis & Schwartz, 8 in Cat. of Life…): Animals, Fungi, Plants : 3 kingdoms Protists : 1 (or 2 if Stramenopiles [Heterokonts] recognized, = Cavalier-Smith’s Kingdom “Chromista”) Bacteria + Archaea : 2 (=1 in Margulis & Schwartz) Viruses : 1 (not in Margulis & Schwartz)](https://image.slidesharecdn.com/rees-irmng-oct-2011-111110170043-phpapp02/75/Tony-Rees-Towards-a-Hierarchical-Classification-of-All-Life-51-2048.jpg)