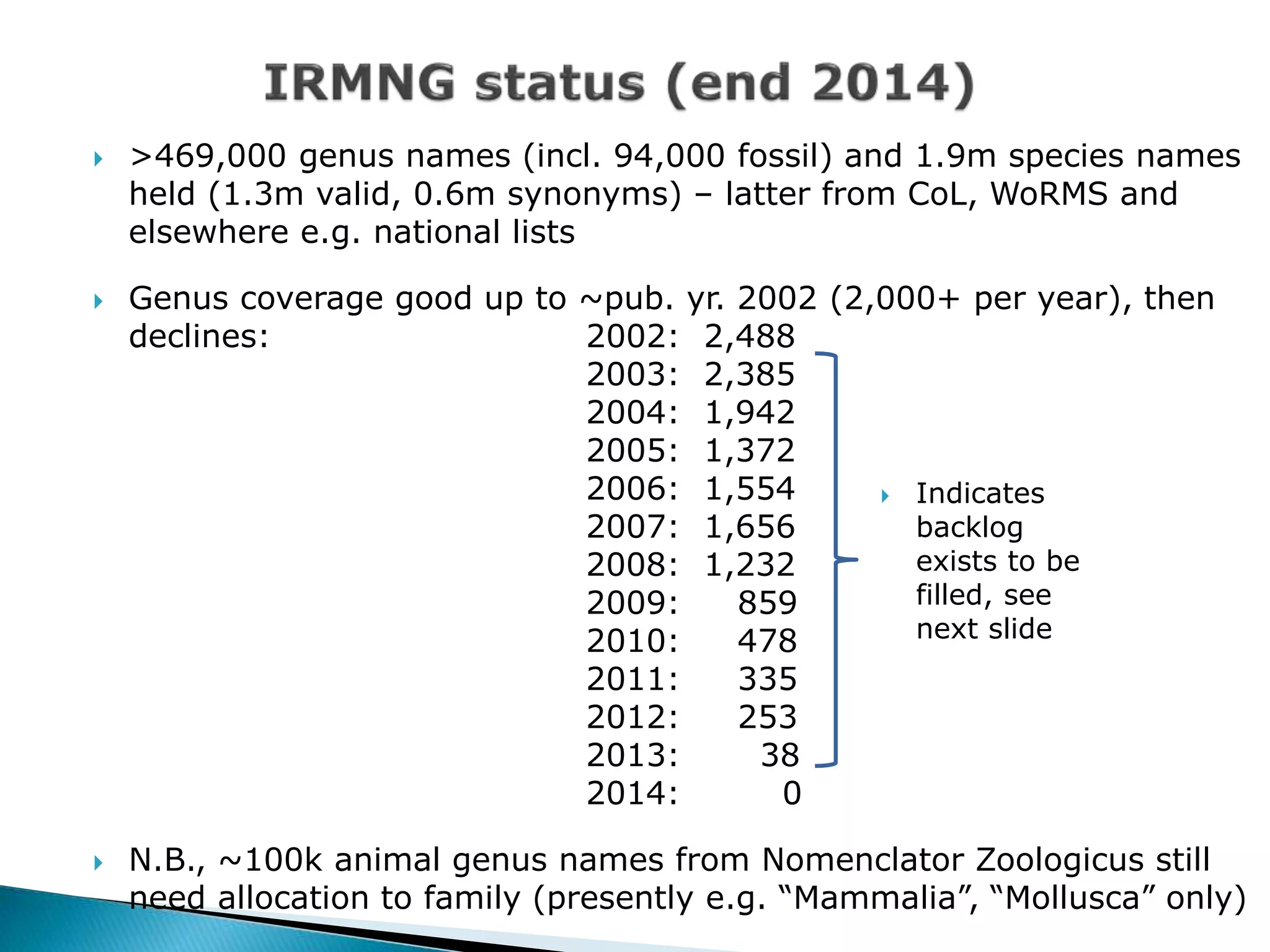
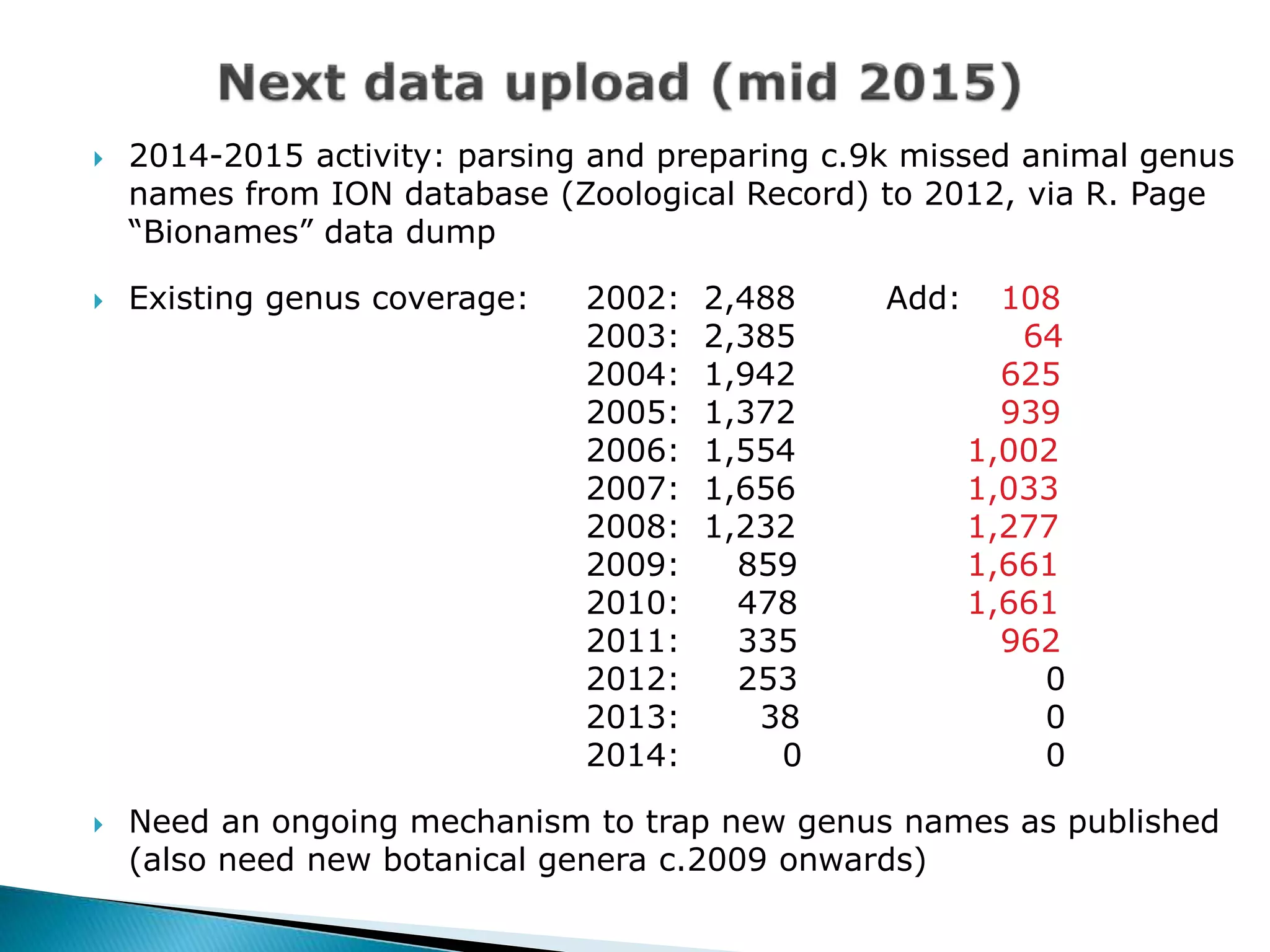
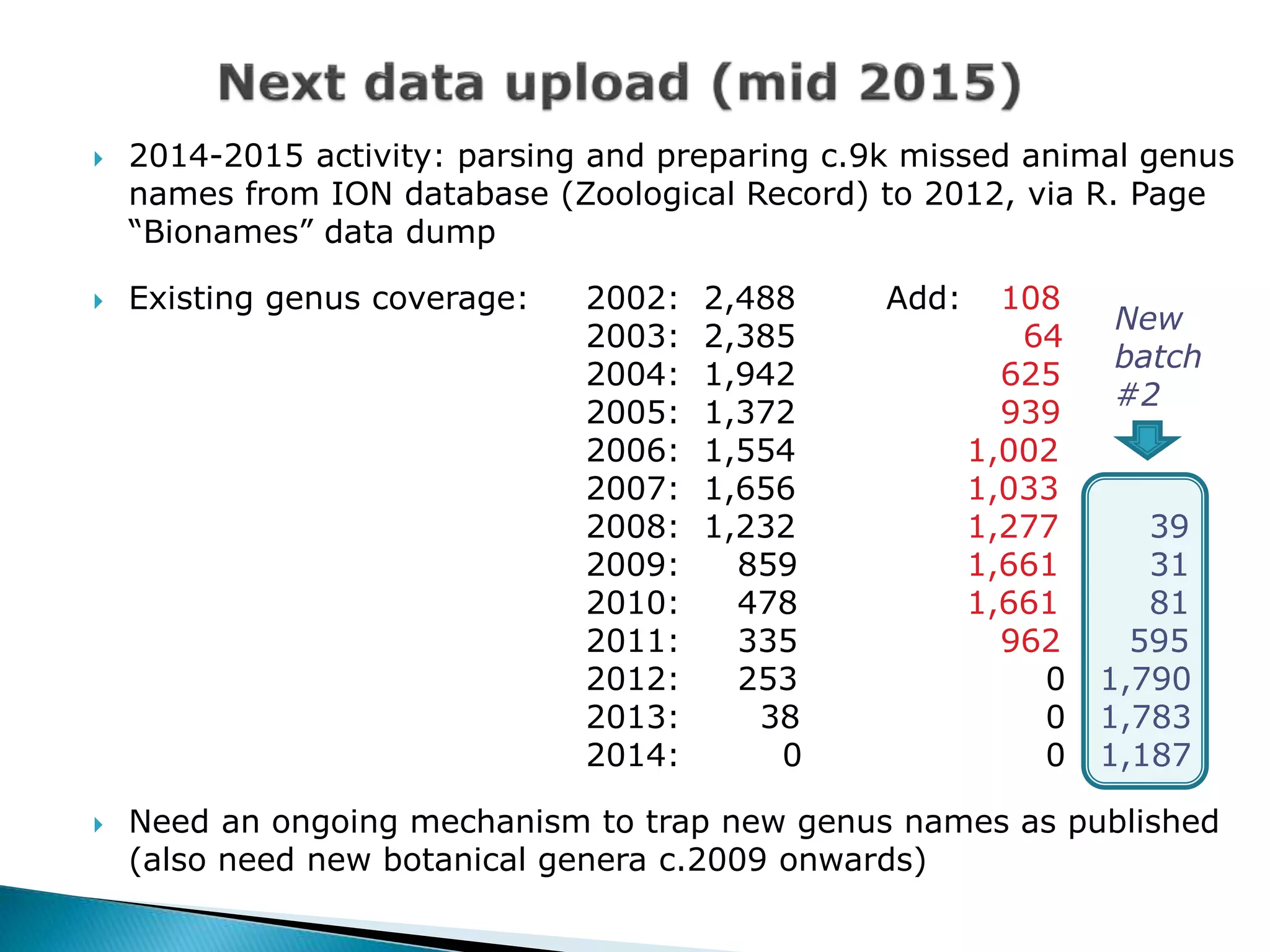
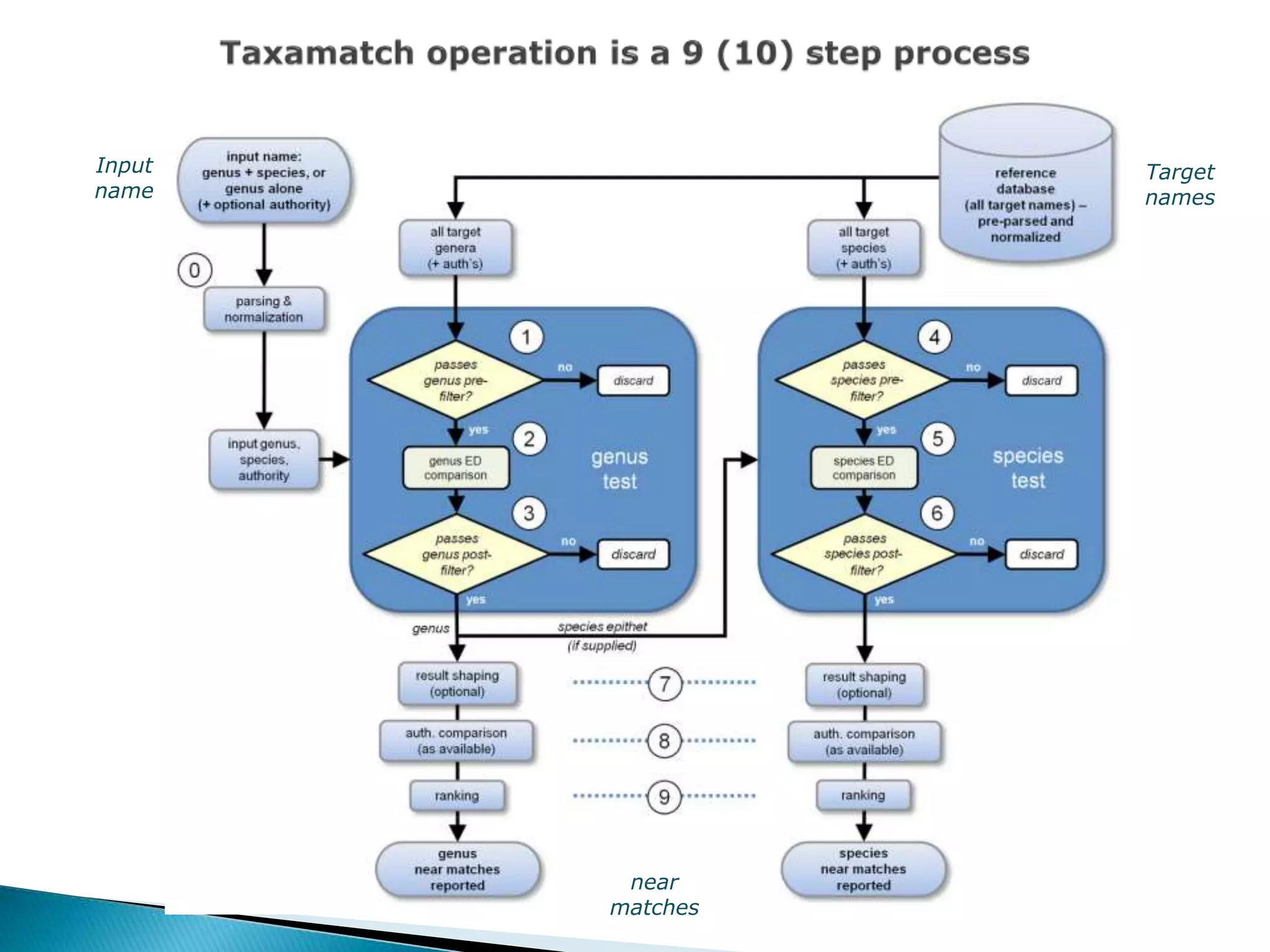
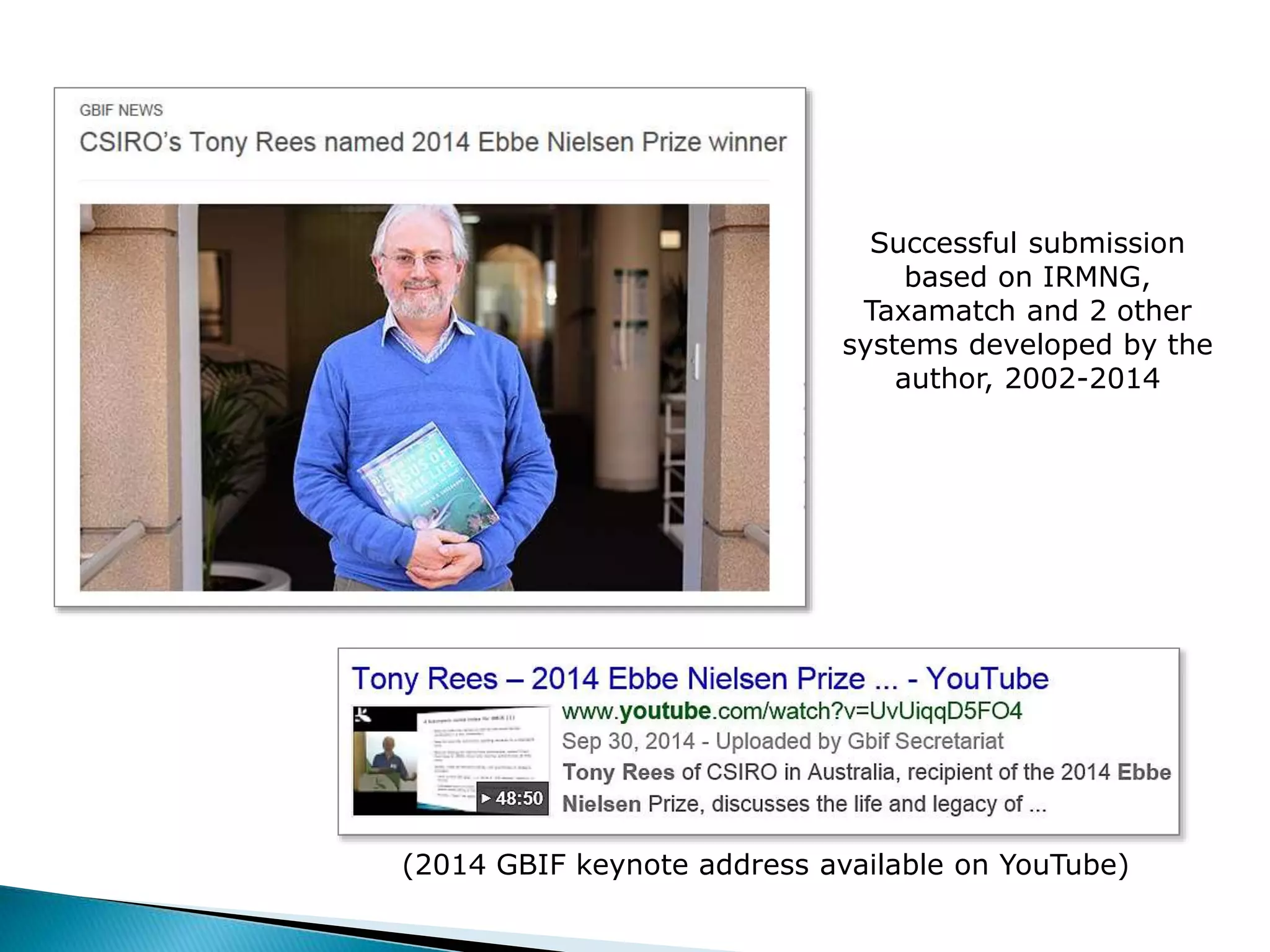
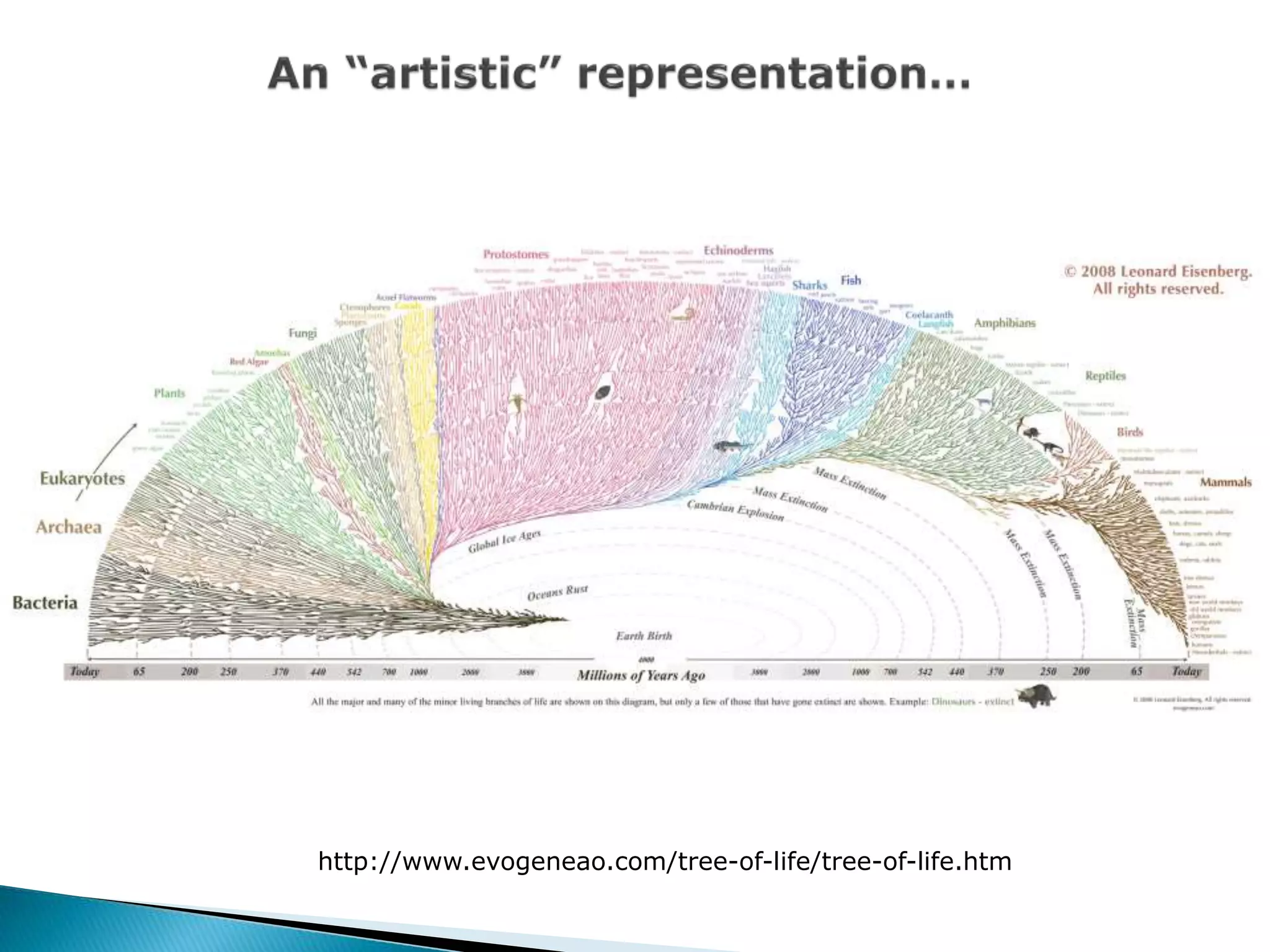
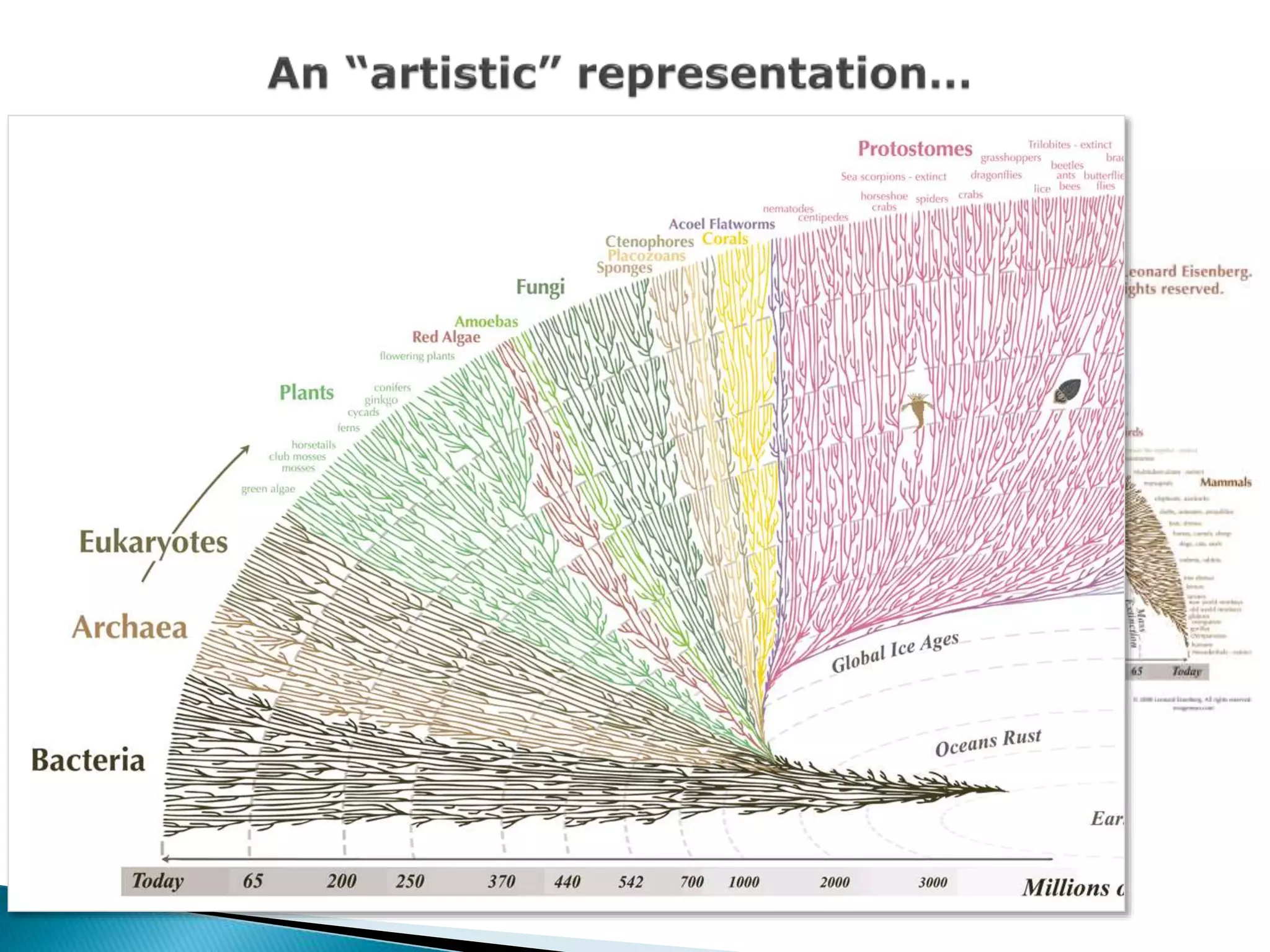
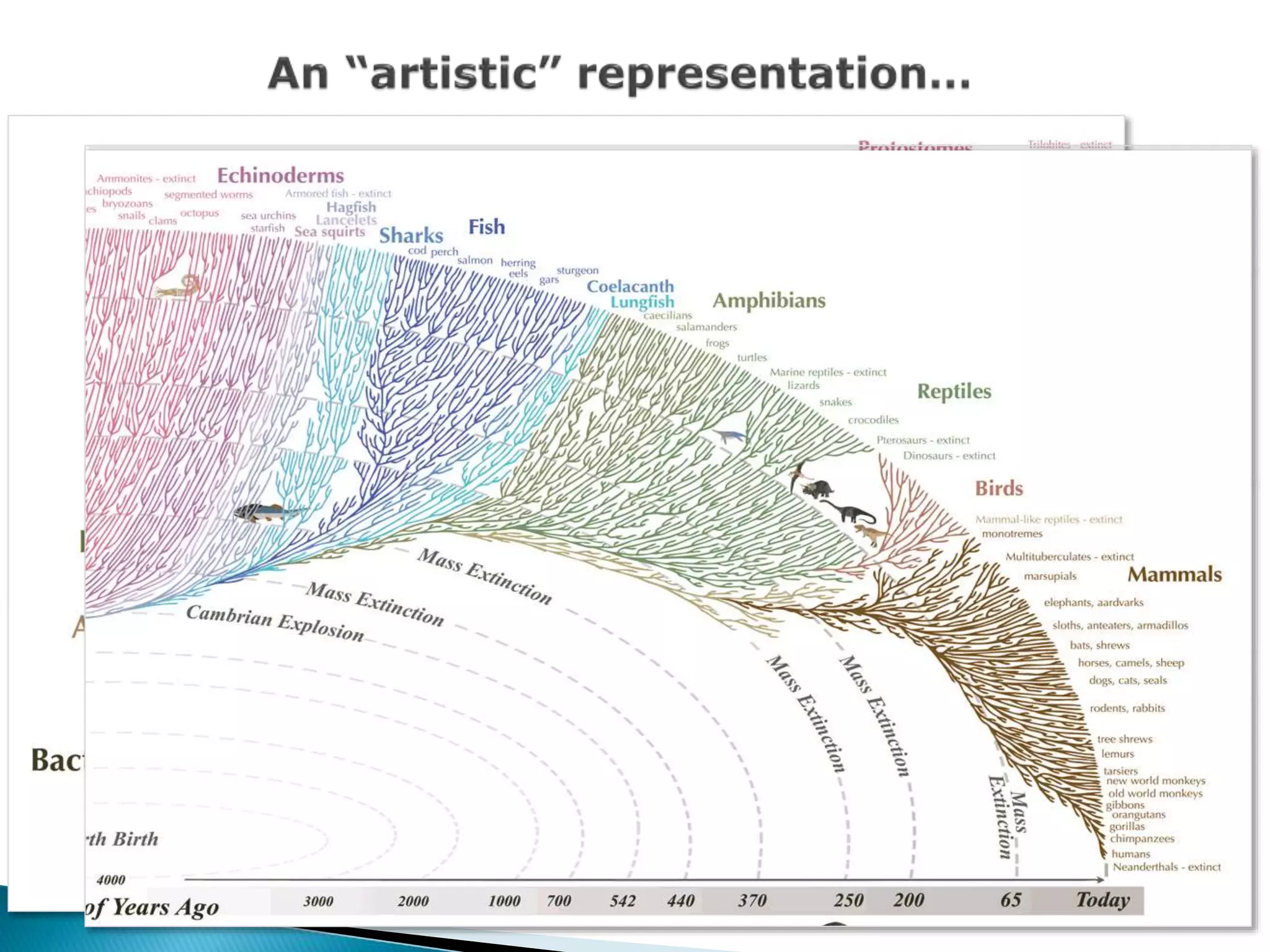
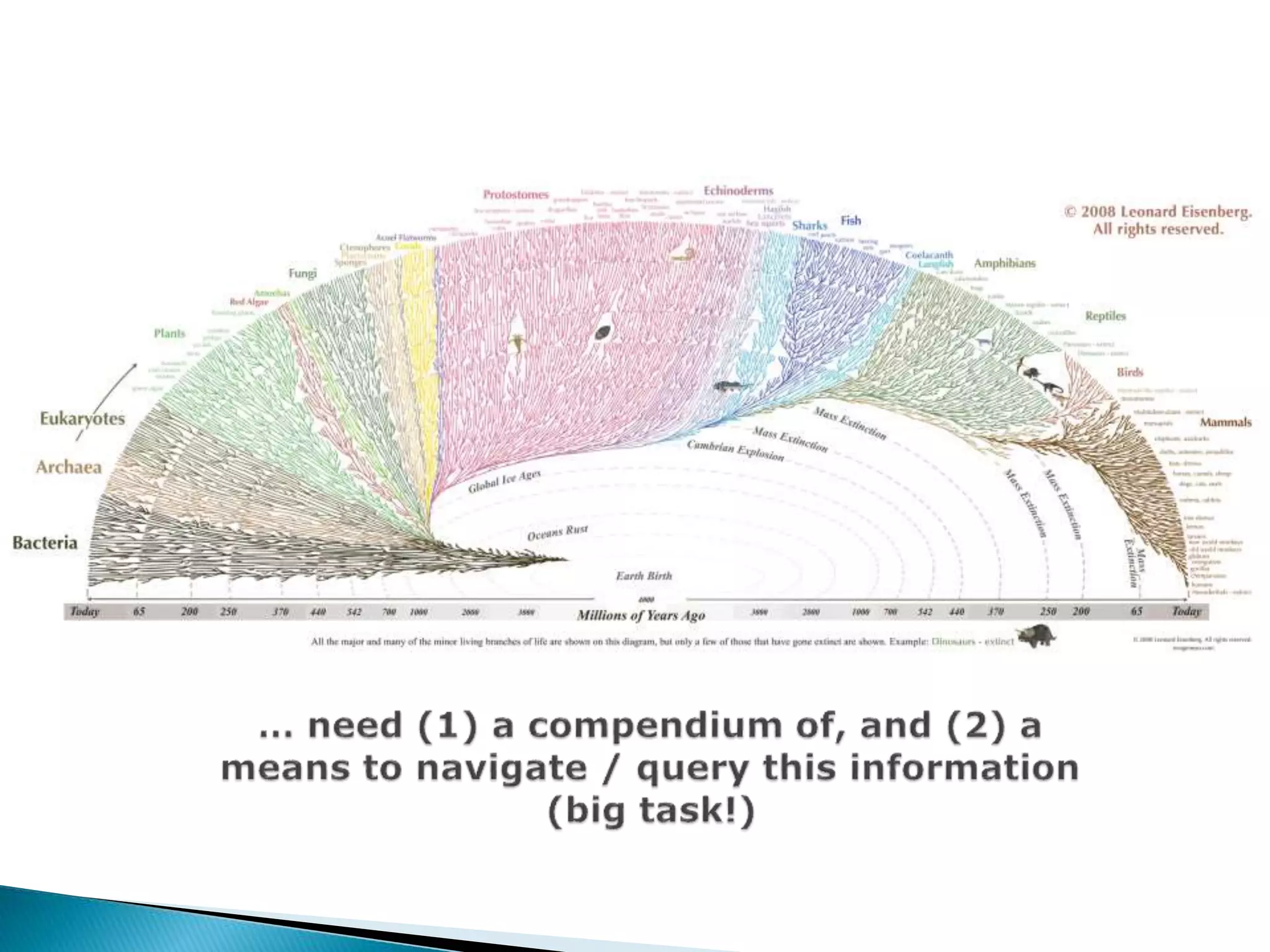
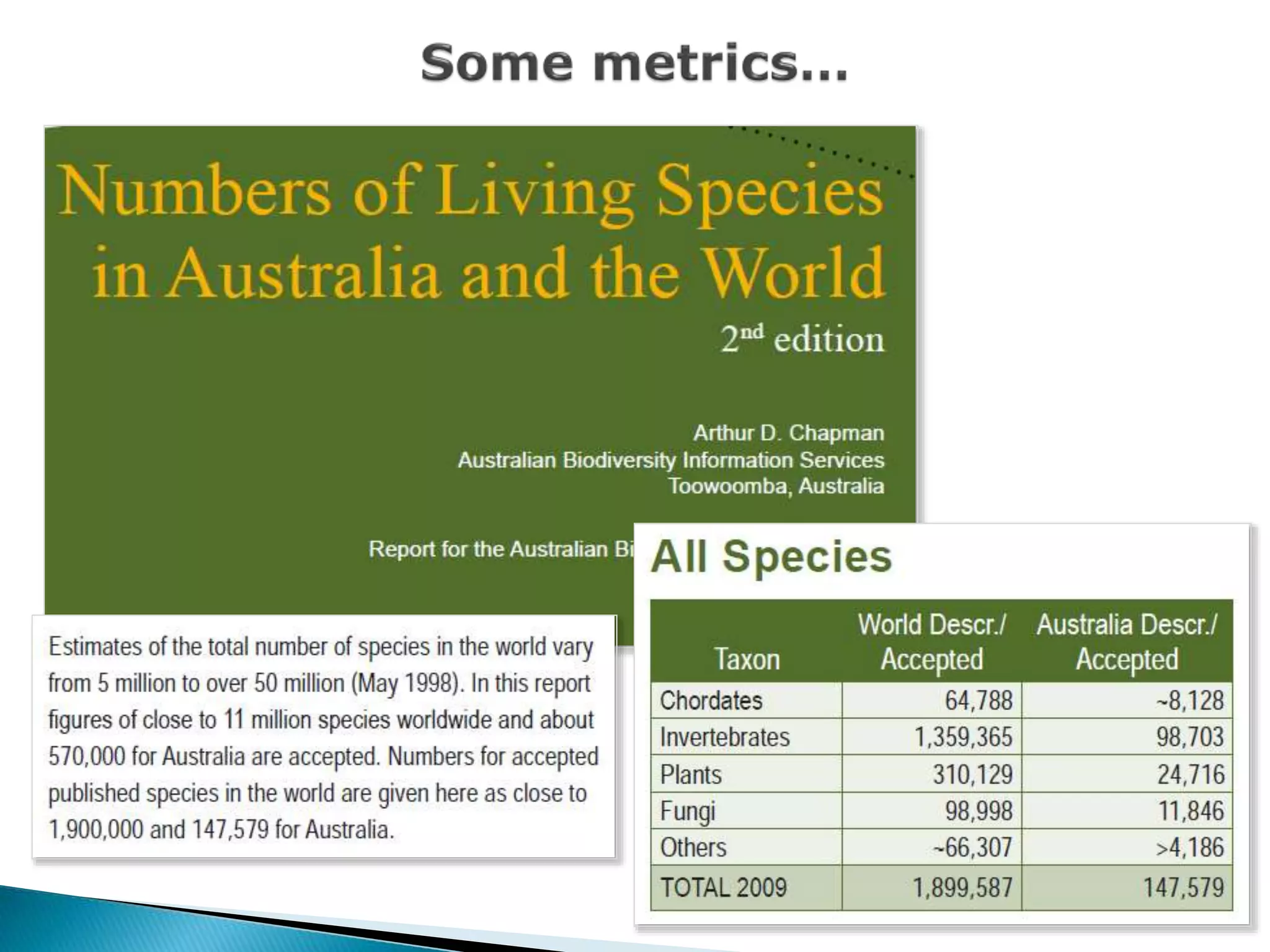
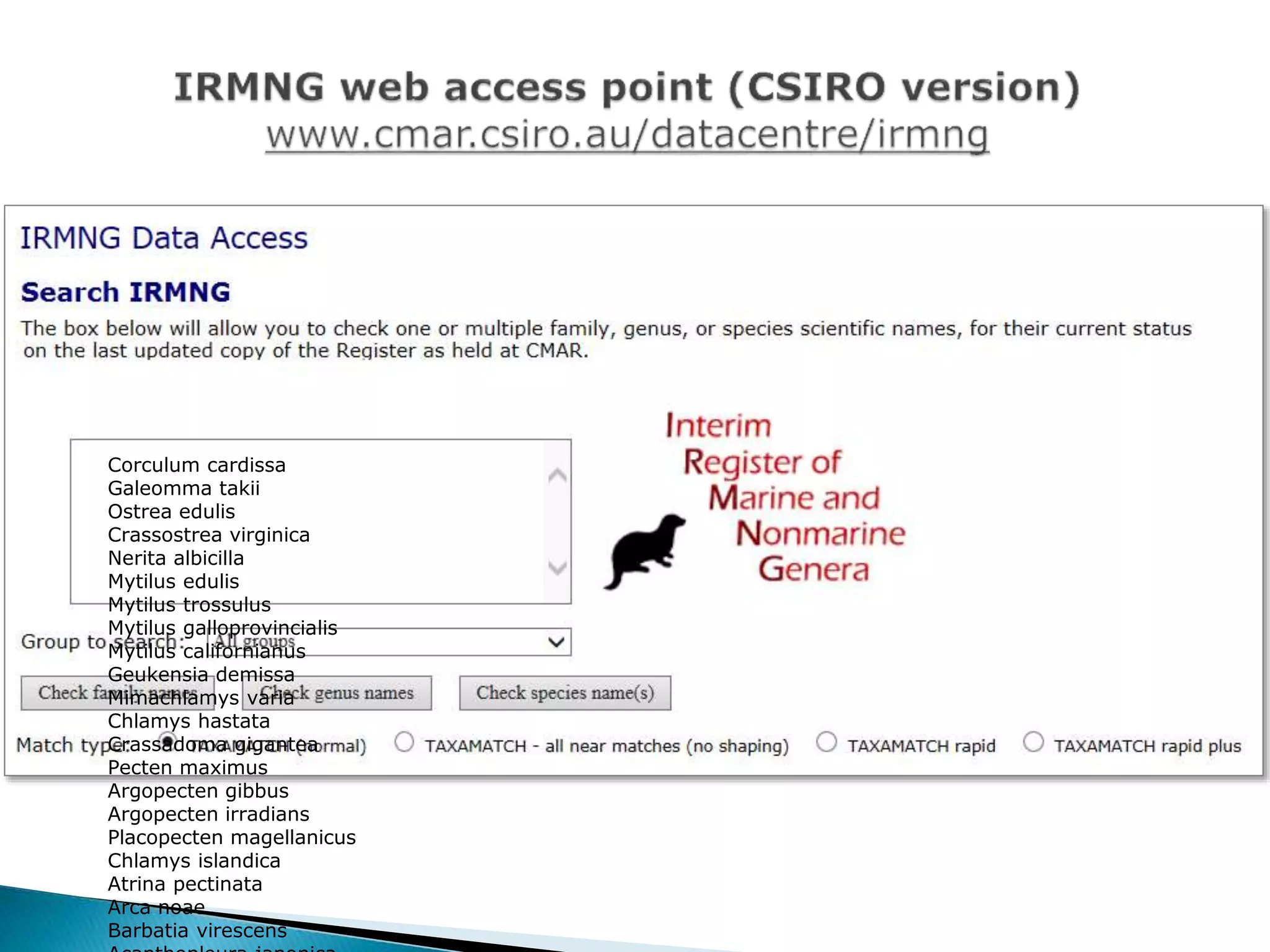
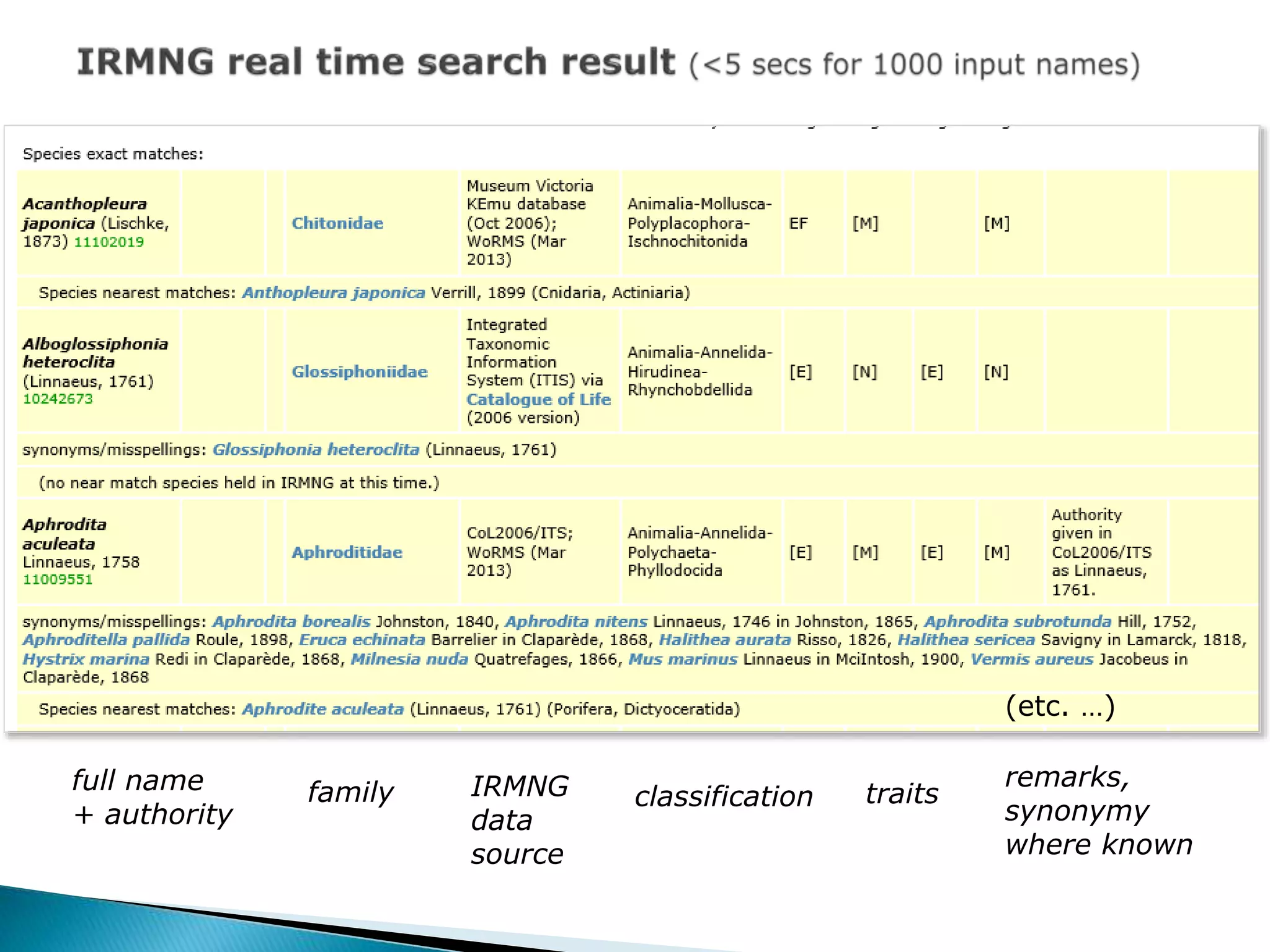
The document addresses bioinformatics and biodiversity informatics, focusing on the digital representation and management of species and biodiversity data. It discusses the challenges in tracking species taxonomy, including synonyms, extinct and extant taxa, and various databases aimed at cataloging this information. The document highlights ongoing efforts to create comprehensive surveys of biodiversity and the importance of collaboration among various databases and projects.





















![ “I've never seen someone's chin hit the floor so quickly when I
showed them IRMNG. I have been asked to show everyone in
the WAM [West Australian Museum] about it, and send out an
email to let them know it is there. At the morning tea it was
discussed and there was a good level of excitement… Part of
this work is reviewing taxonomic names in the Kimberley. We
are going to keep an eye out for any names that we identify as
errors and can feed that back.”
– Piers Higgs, Gaia Resources, Australia
[Piers@gaiaresources.com.au], 21 May 2009](https://image.slidesharecdn.com/rees-irmng-2015-150831060915-lva1-app6891/75/Tony-Rees-IRMNG-2015-presentation-39-2048.jpg)
![ “I've never seen someone's chin hit the floor so quickly when I
showed them IRMNG. I have been asked to show everyone in
the WAM [West Australian Museum] about it, and send out an
email to let them know it is there. At the morning tea it was
discussed and there was a good level of excitement… Part of
this work is reviewing taxonomic names in the Kimberley. We
are going to keep an eye out for any names that we identify as
errors and can feed that back.”
– Piers Higgs, Gaia Resources, Australia
[Piers@gaiaresources.com.au], 21 May 2009
“IRMNG is the most useful web tool I've ever used - I also used
it to place Micropalaeontology genera into families (ostracods
and foraminifera) - Here there is always a single match in these
groups, sometimes also a match in Mollusca and Arthropoda,
but I just use the former ones because I know that these
names are of specimens belonging to those groups… Thank you
very much for developing this fantastic tool.”–
– Willem Coetzer, South African Institute for Aquatic
Biodiversity [w.coetzer@saiab.ac.za], 2 April 2011](https://image.slidesharecdn.com/rees-irmng-2015-150831060915-lva1-app6891/75/Tony-Rees-IRMNG-2015-presentation-40-2048.jpg)