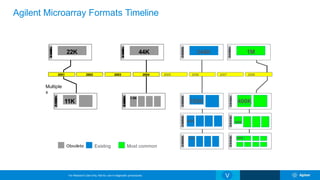
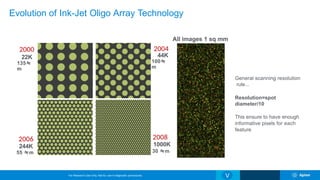
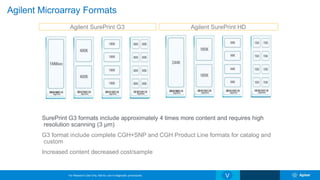
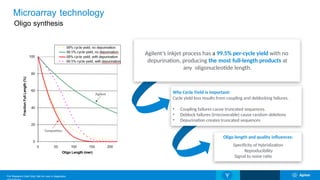
The Agilent microarray platform is designed for research purposes, providing advanced tools for hybridization and sample analysis without diagnostic applications. It features a variety of formats and customization capabilities, emphasizing the importance of oligo length and quality for sensitivity and specificity in copy number detection. The technology allows for detailed gene expression analysis and integration with next-generation sequencing for enhanced data interpretation.