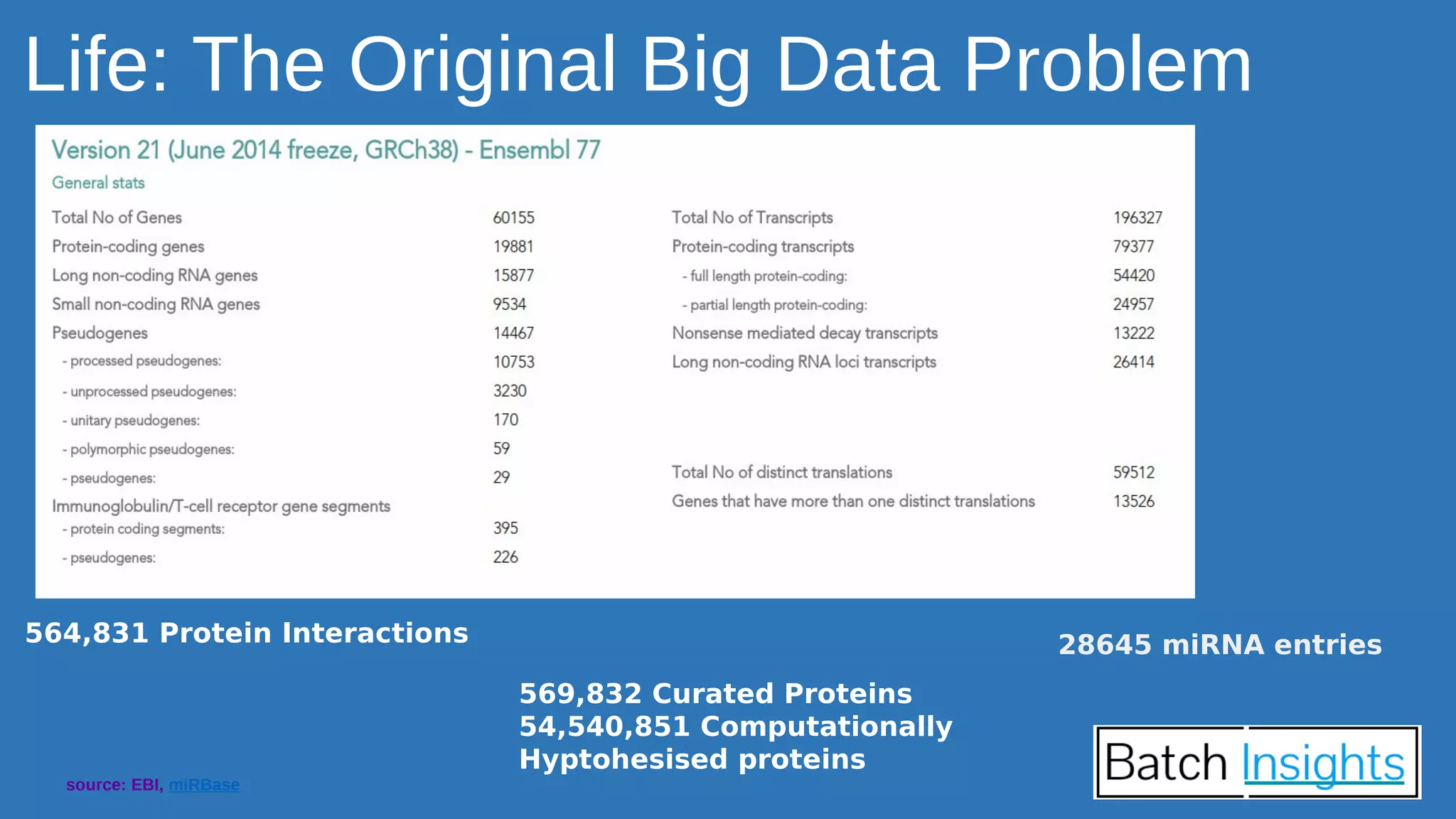
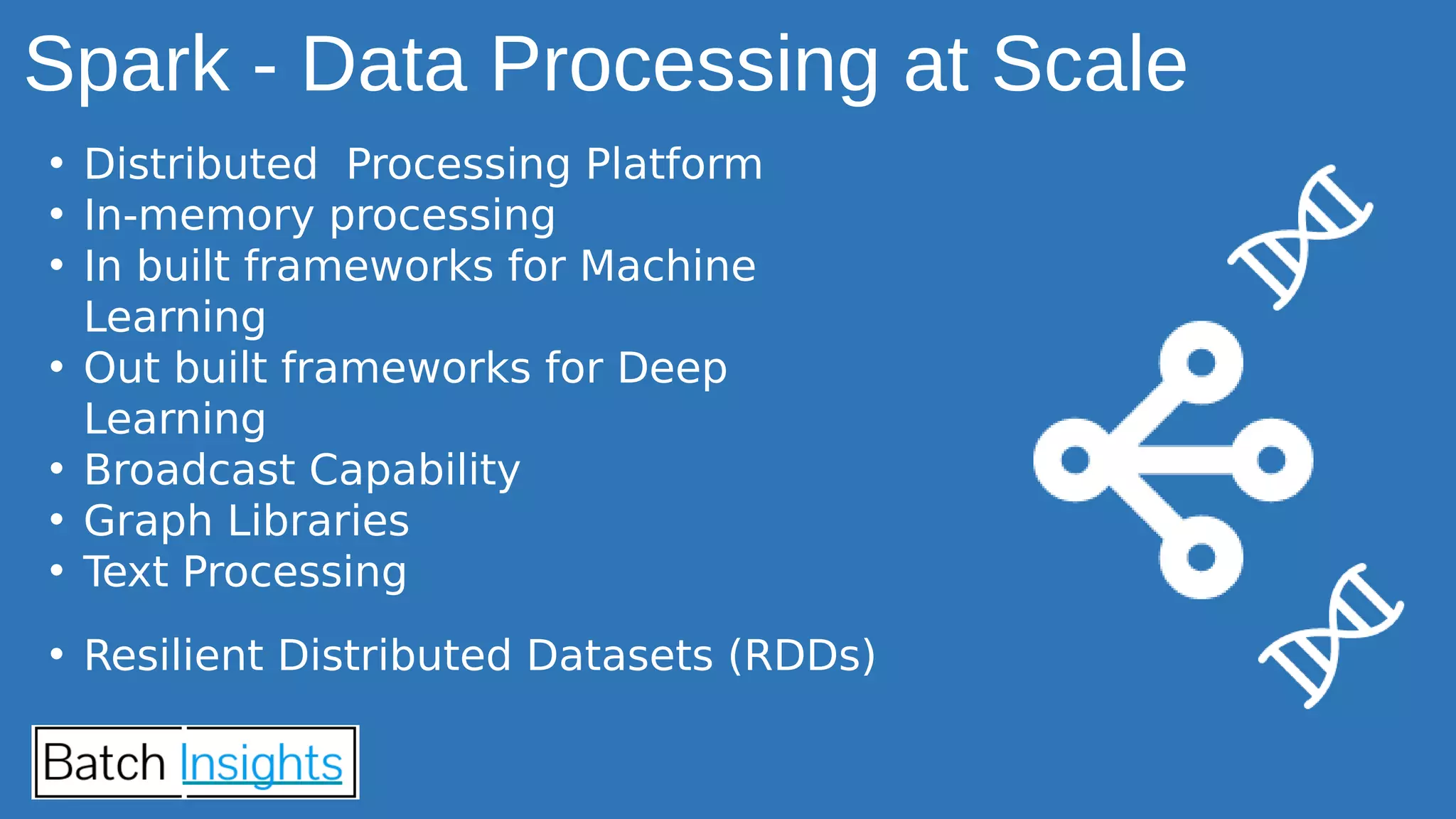
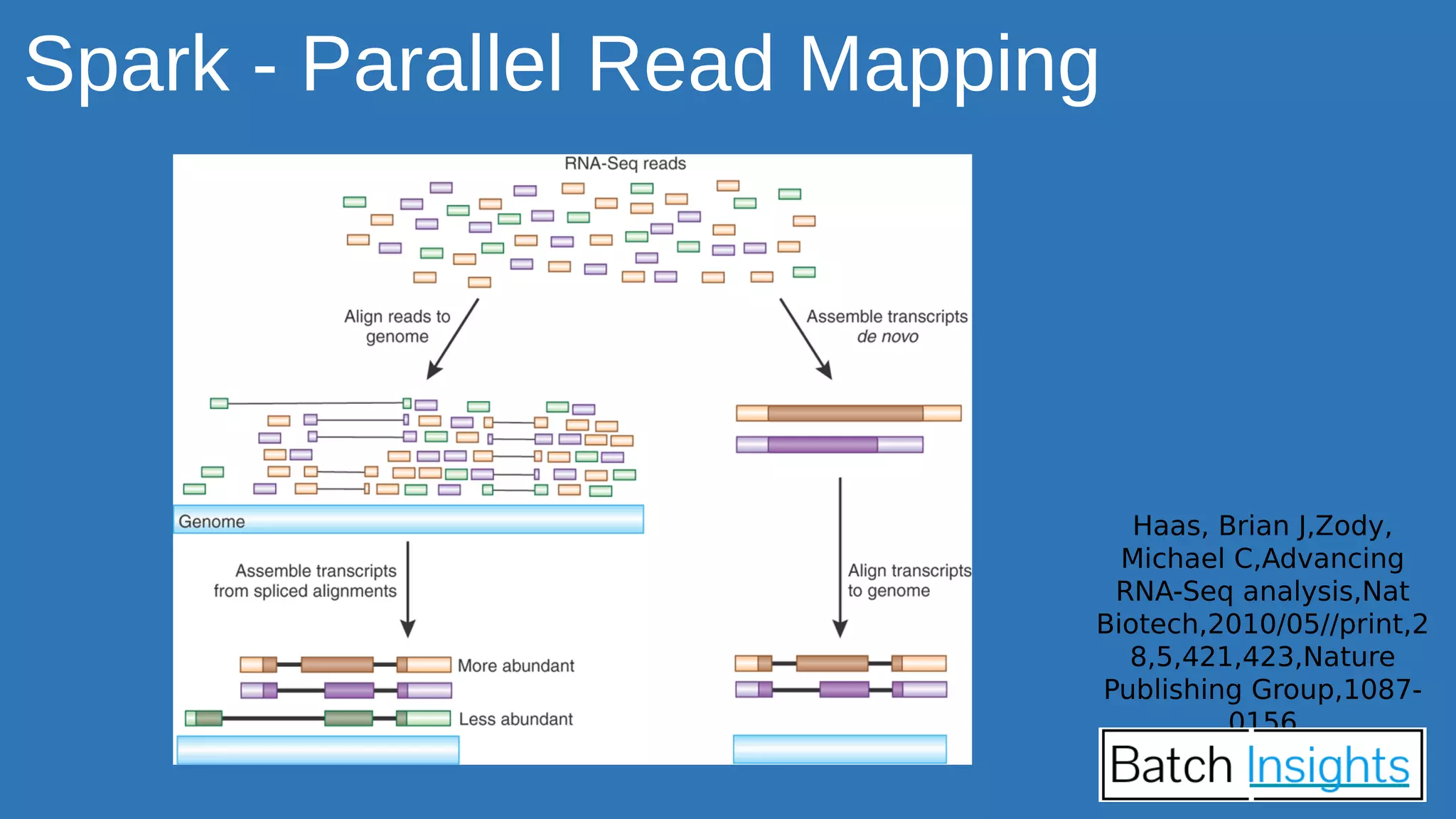
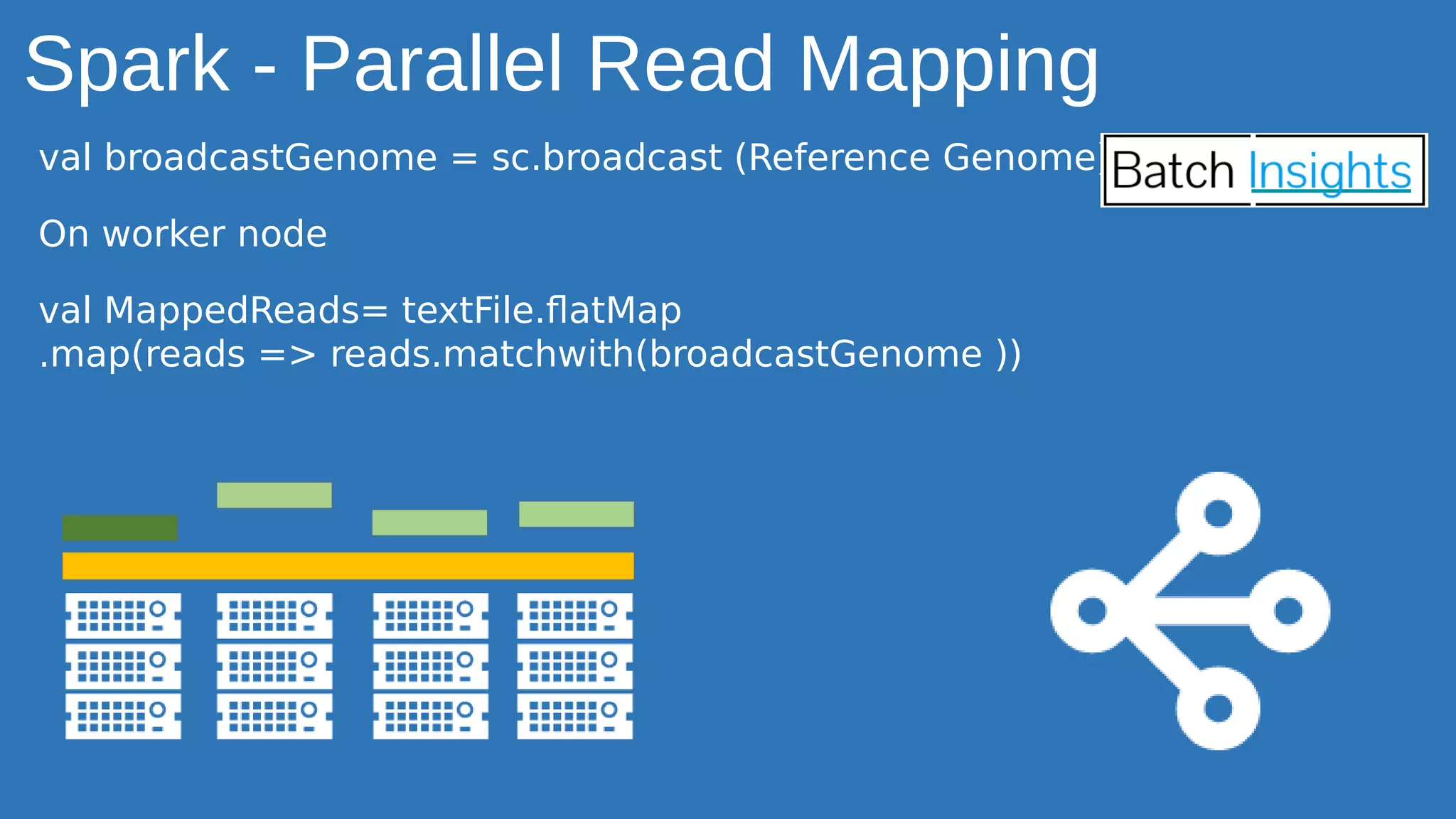
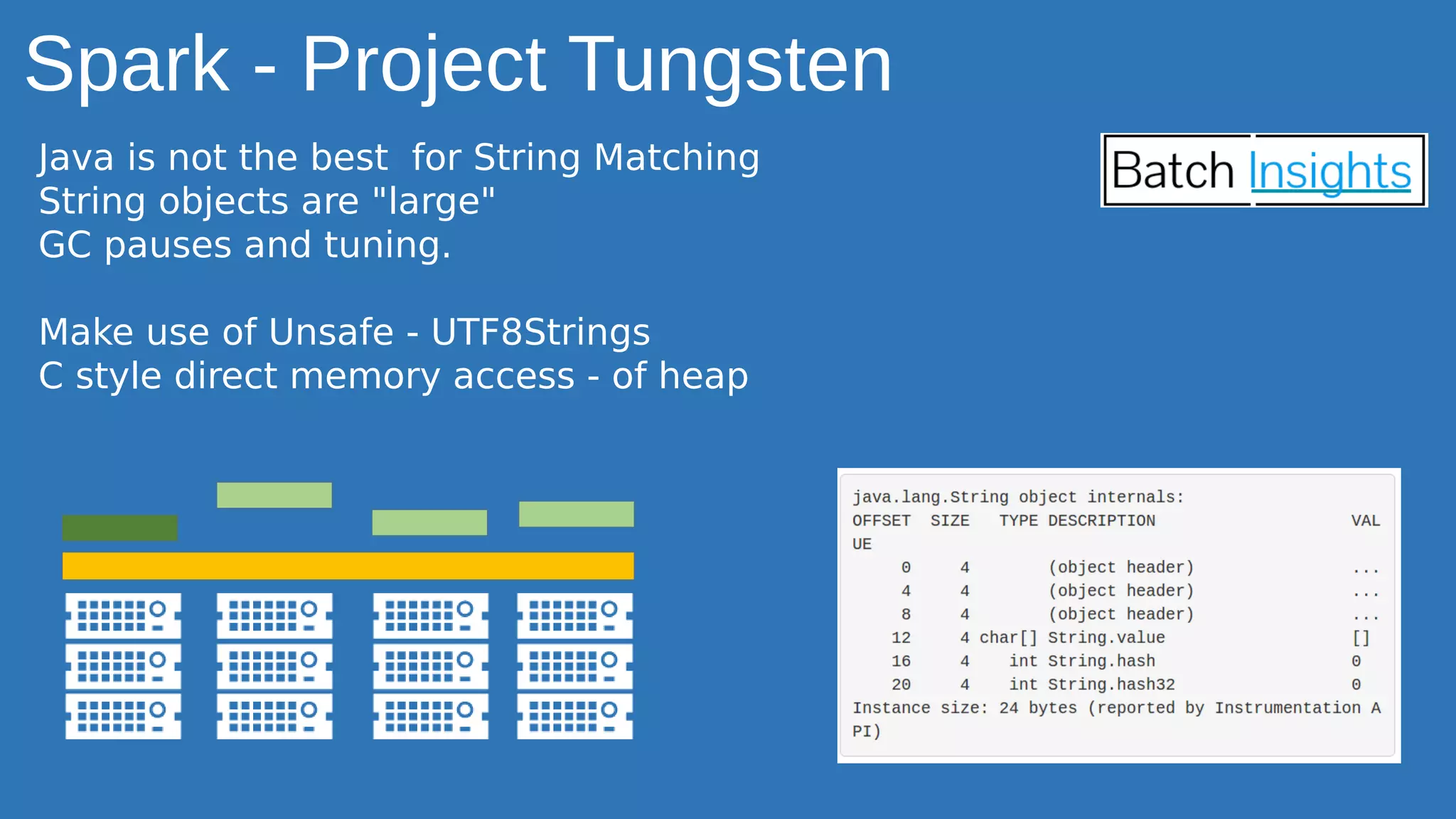
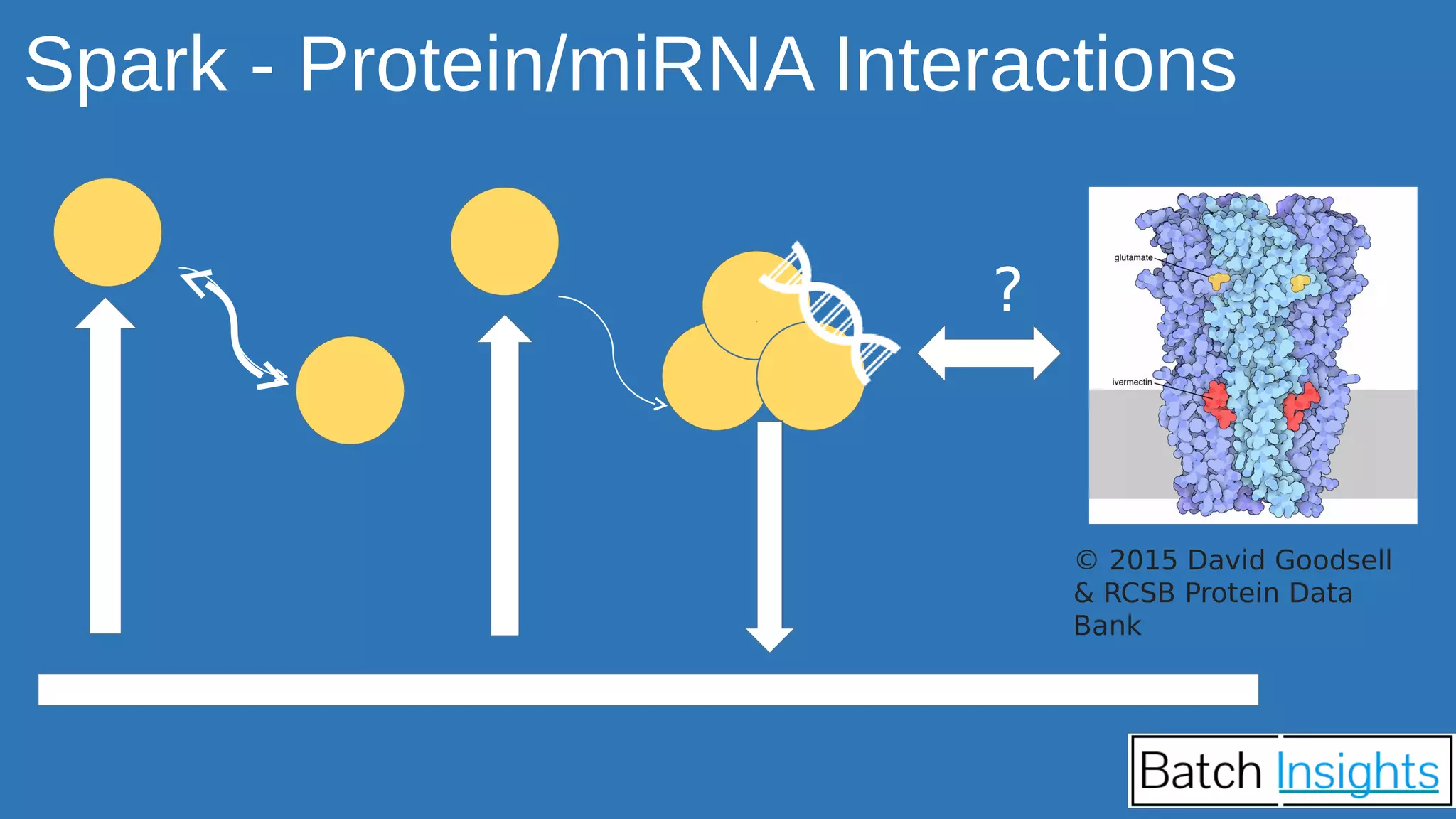
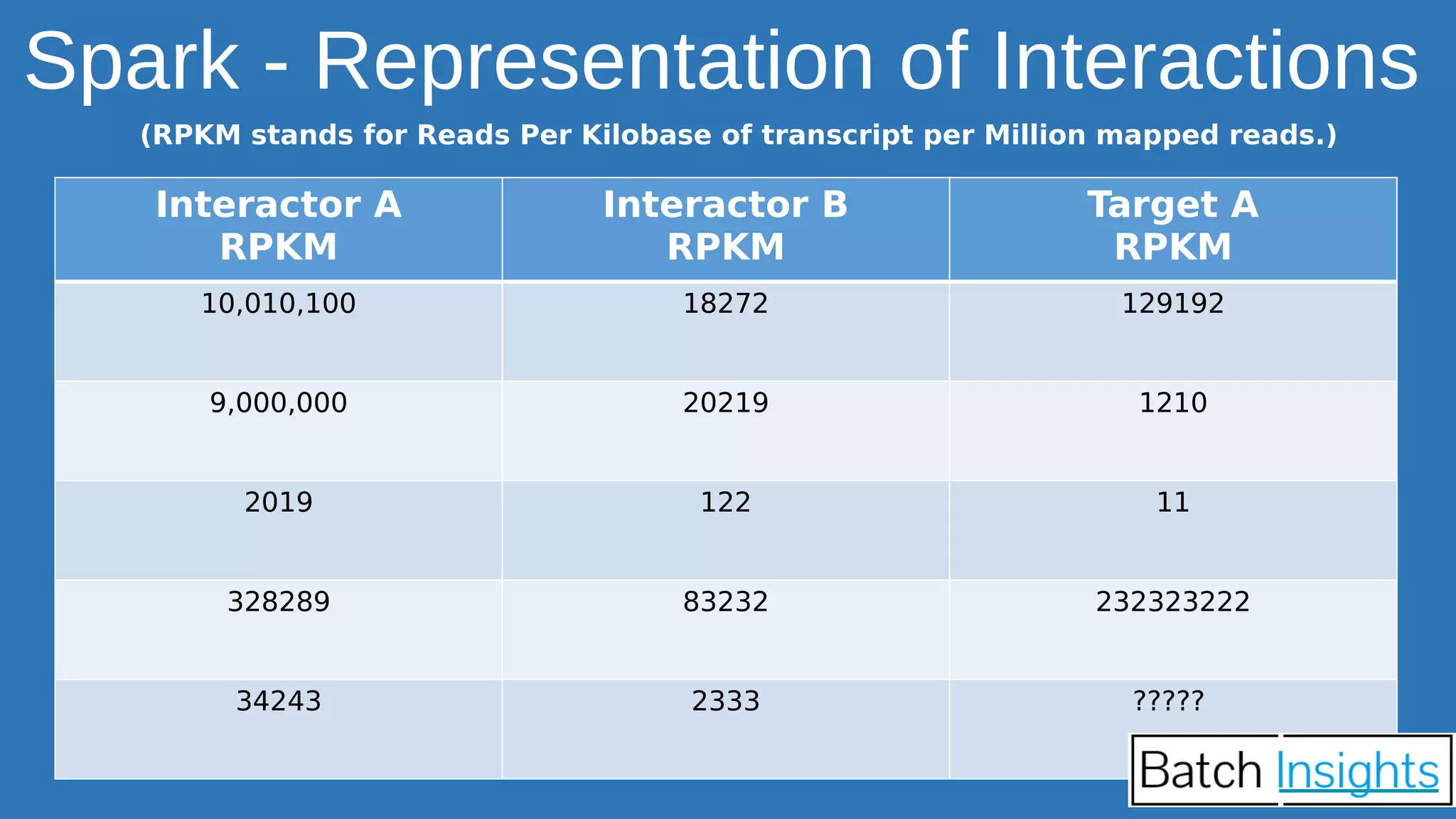
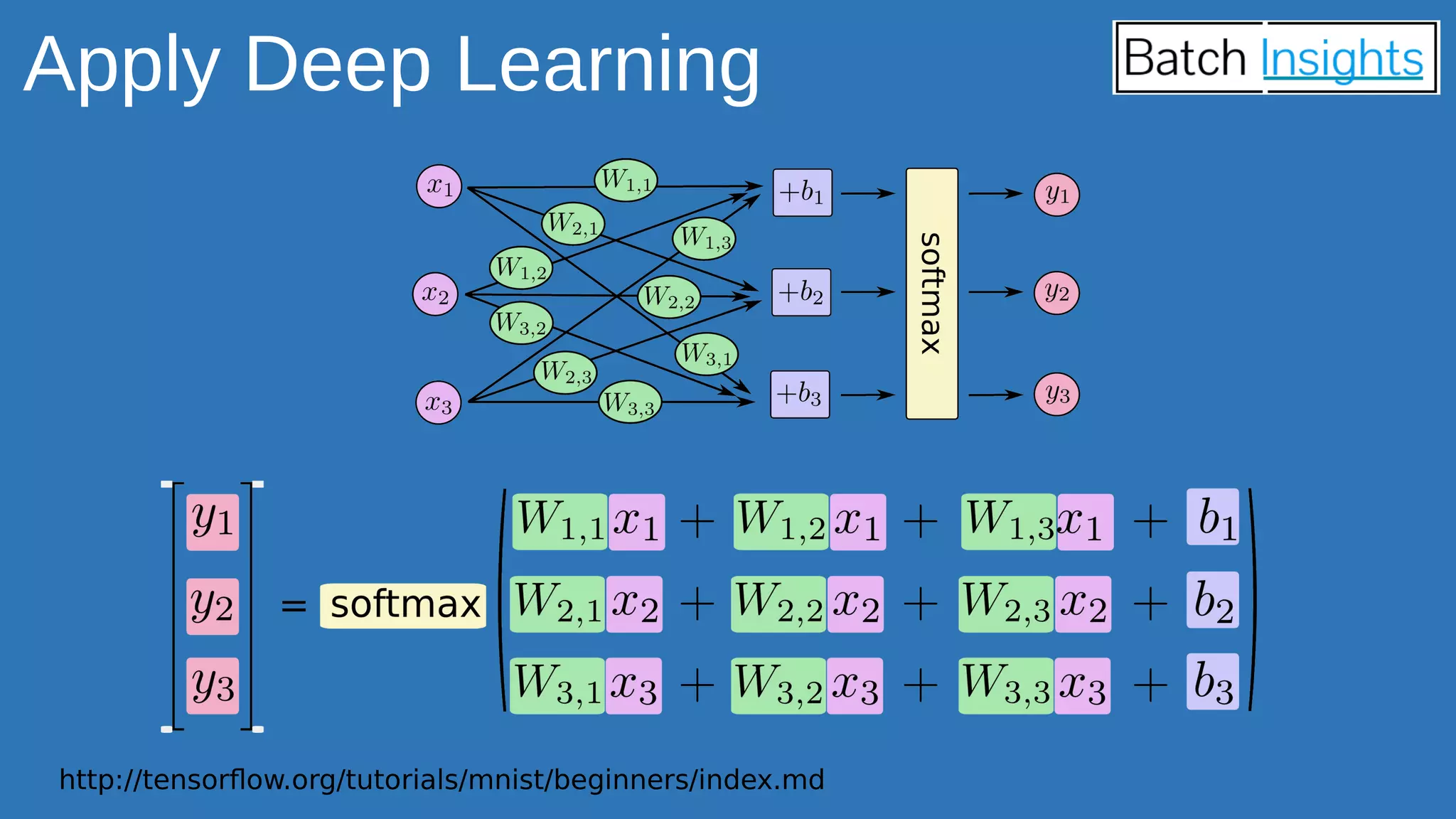
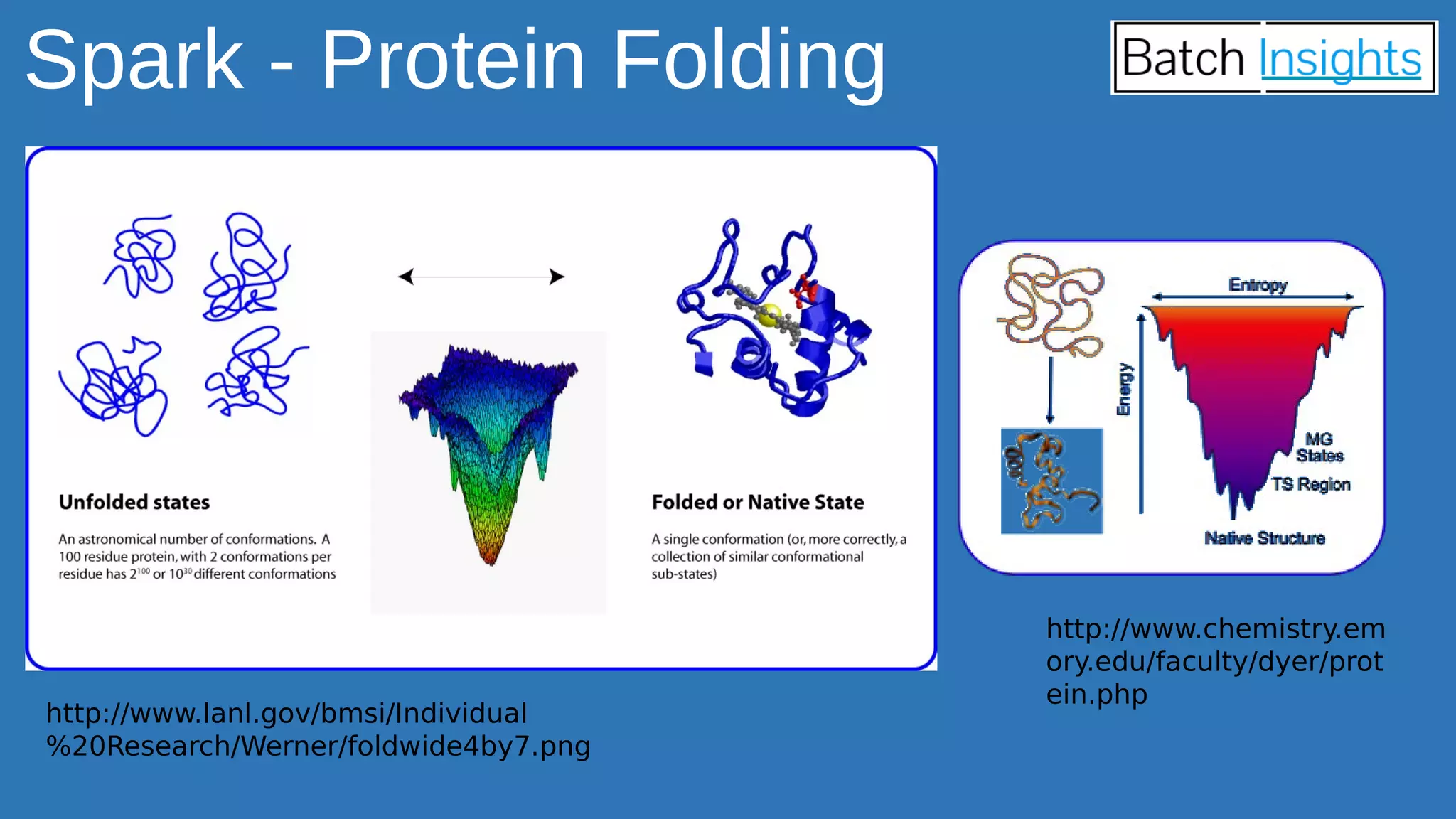
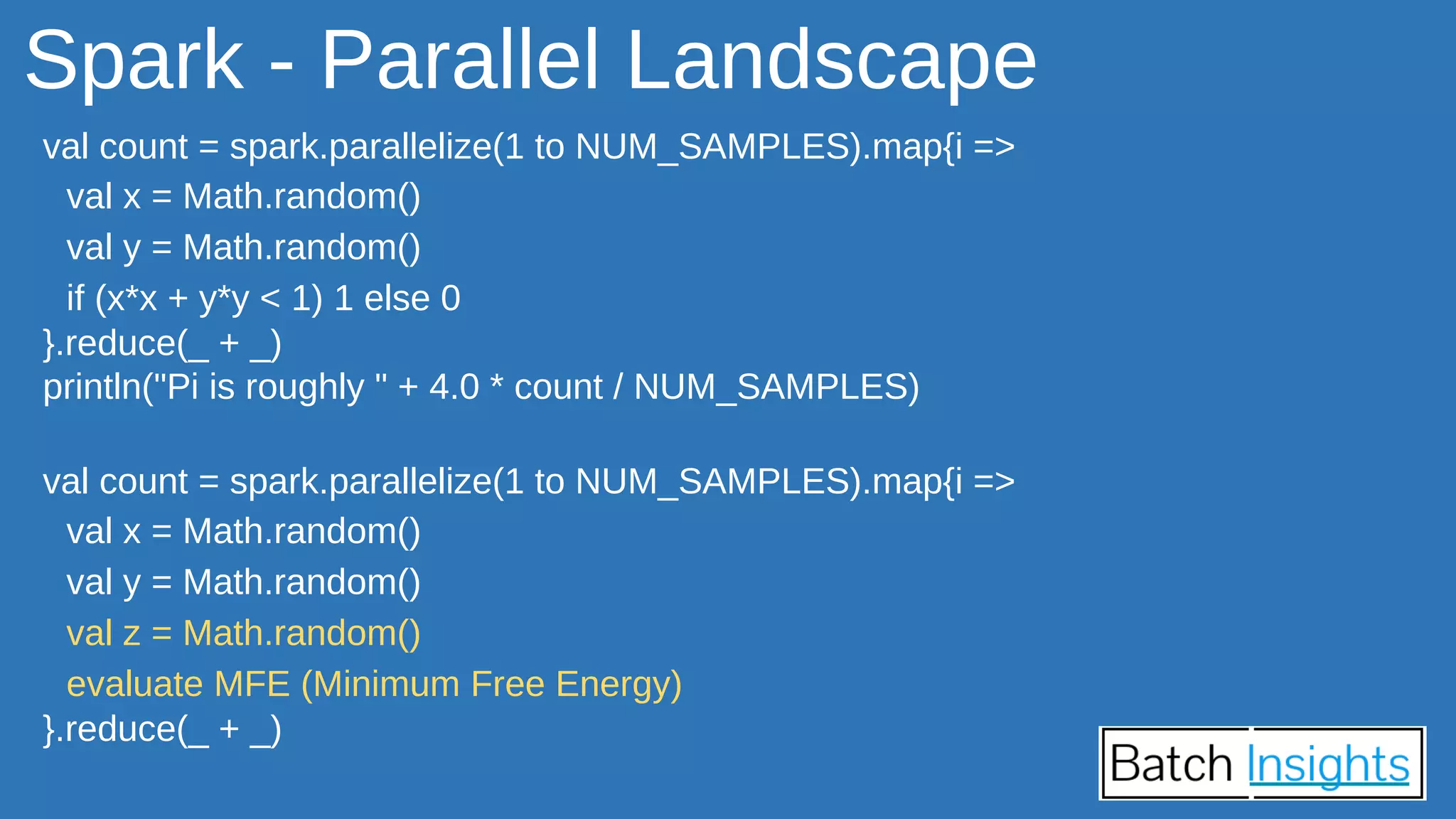
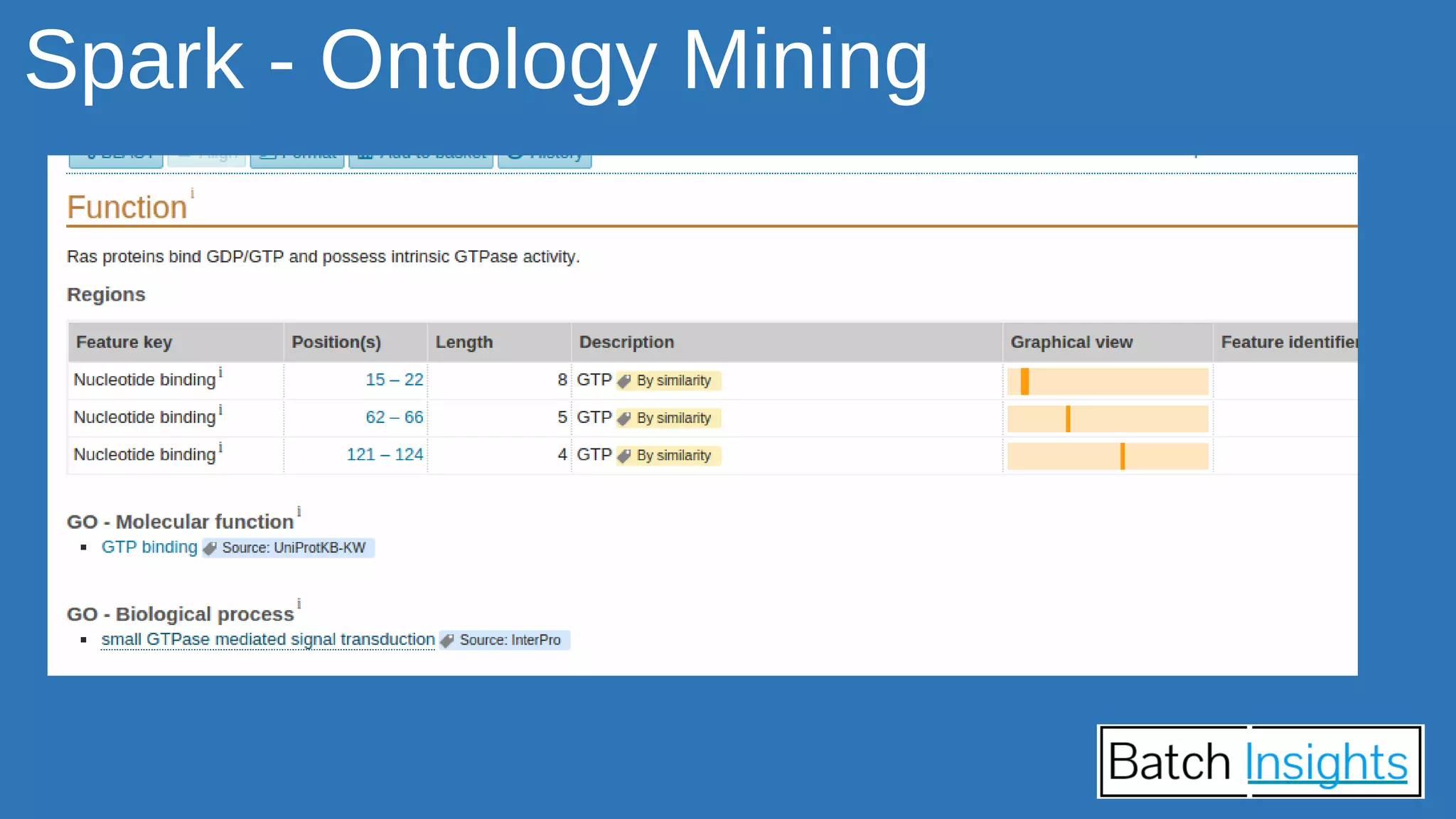
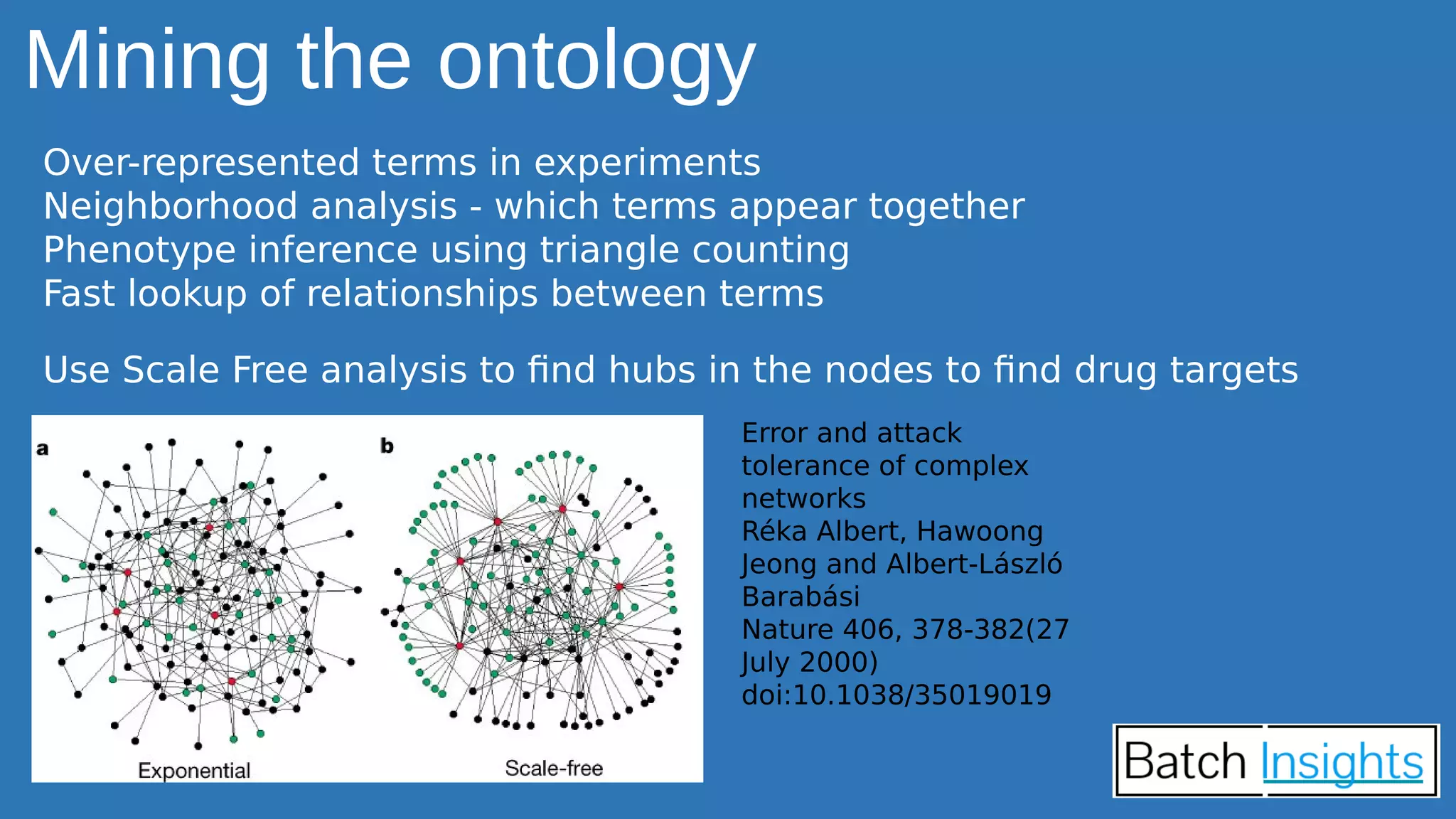
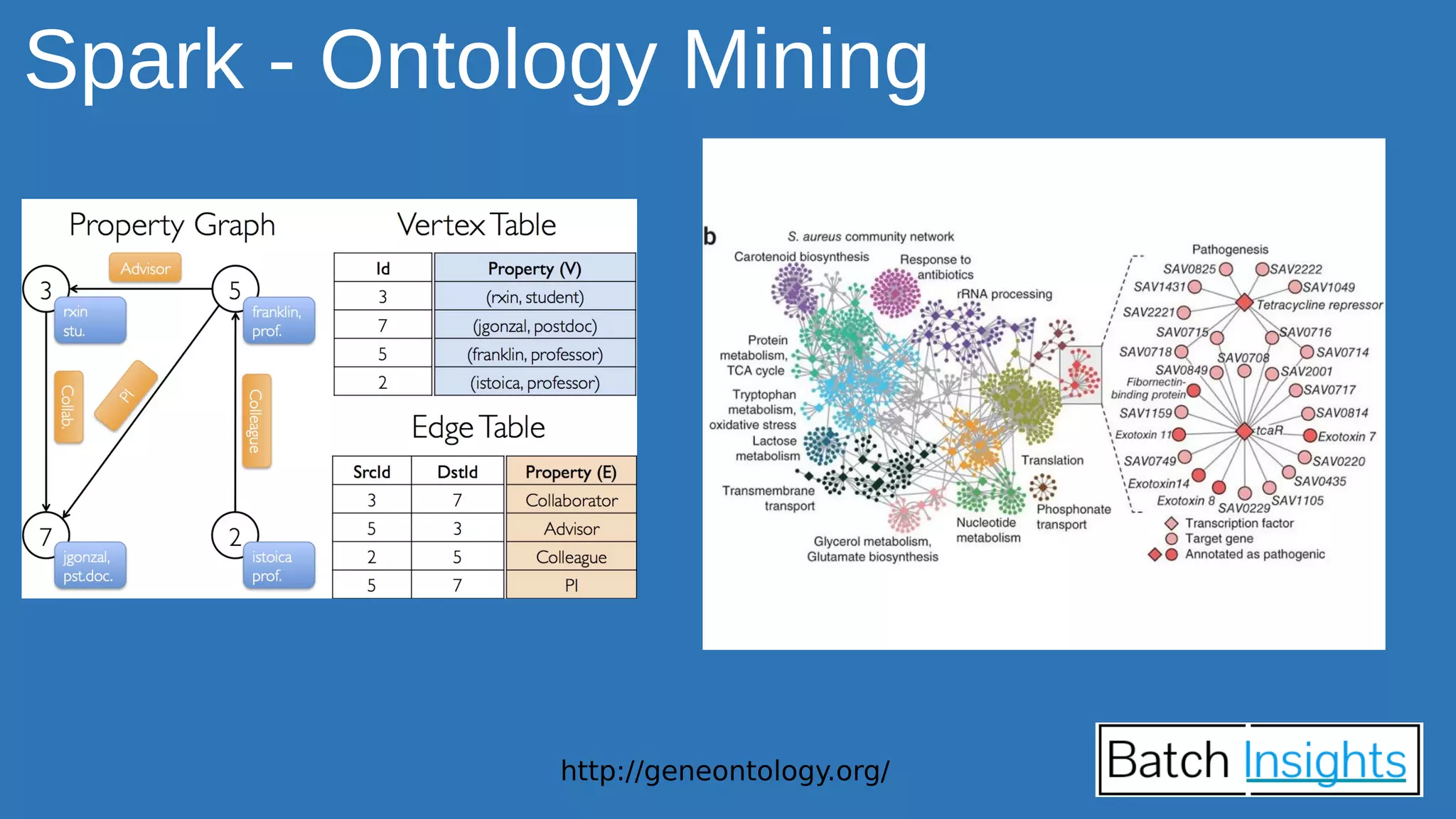
The document discusses the applications of Apache Spark in life sciences, particularly in systems biology and deep learning for protein interaction and folding analysis. It highlights the challenges of processing large datasets and emphasizes the use of distributed and in-memory processing frameworks for tasks such as RNA-seq analysis and ontology mining. Additionally, it outlines various use cases, including virtual drug screening and disease modeling.