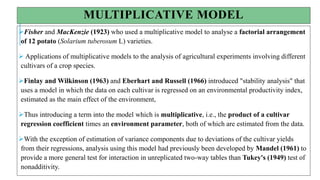
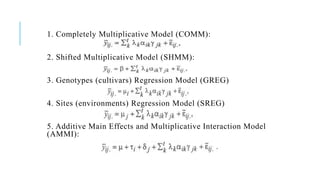
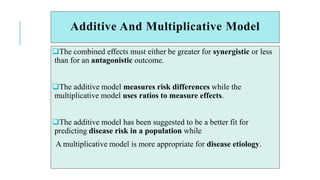
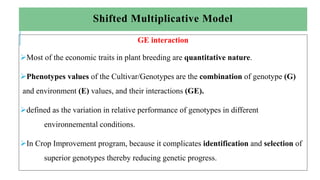
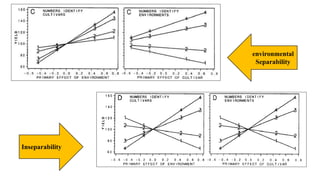
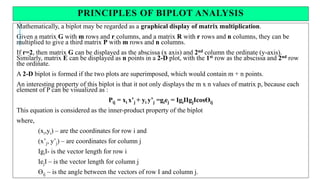
The document covers advanced topics in biometrical and quantitative genetics, specifically discussing various models for analyzing genotype and environmental interactions. Key points include the additive and multiplicative models, the Alternating Conditional Expectations (ACE) algorithm, and the significance of genotype x environment interactions (GEI) in crop improvement. Biplot analysis is highlighted as a valuable tool for visualizing relationships in genotype by environment data, aiding breeders in selecting superior genotypes.