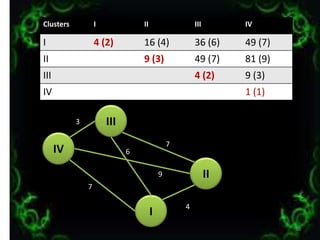
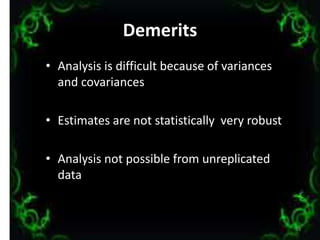
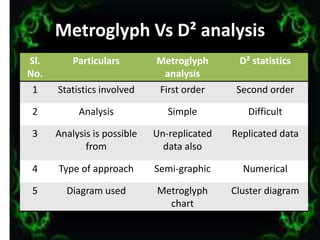
The document discusses genetic diversity analysis, focusing on variability sources, polygenic traits, and methods for assessing genetic diversity, including metroglyph analysis and D2 statistics. It outlines characteristics of phenotypic, genotypic, and environmental variations, along with procedures for evaluating germplasm lines. Additionally, it compares metroglyph and D2 analyses highlighting their respective merits and demerits in studying genetic diversity.