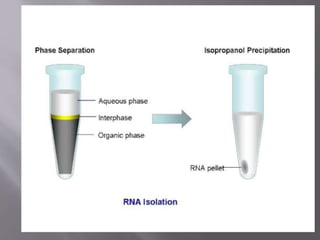
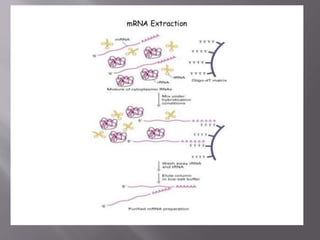
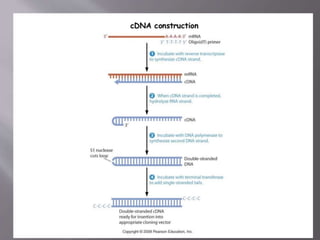
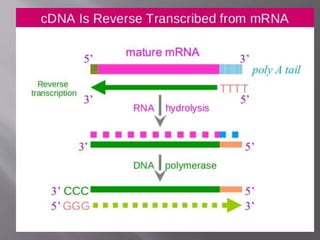
The document outlines the process of RNA isolation using Trizol reagent, highlighting its effectiveness in preserving RNA integrity during tissue homogenization and the separation of RNA from DNA and proteins through chloroform addition. It describes the steps for RNA purification and subsequent cDNA synthesis, including the use of reverse transcriptase and purification methods like column purification to obtain mRNA. Additionally, the document emphasizes the significance of cDNA libraries for gene expression studies in prokaryotic systems, eliminating non-coding regions for efficient analysis.