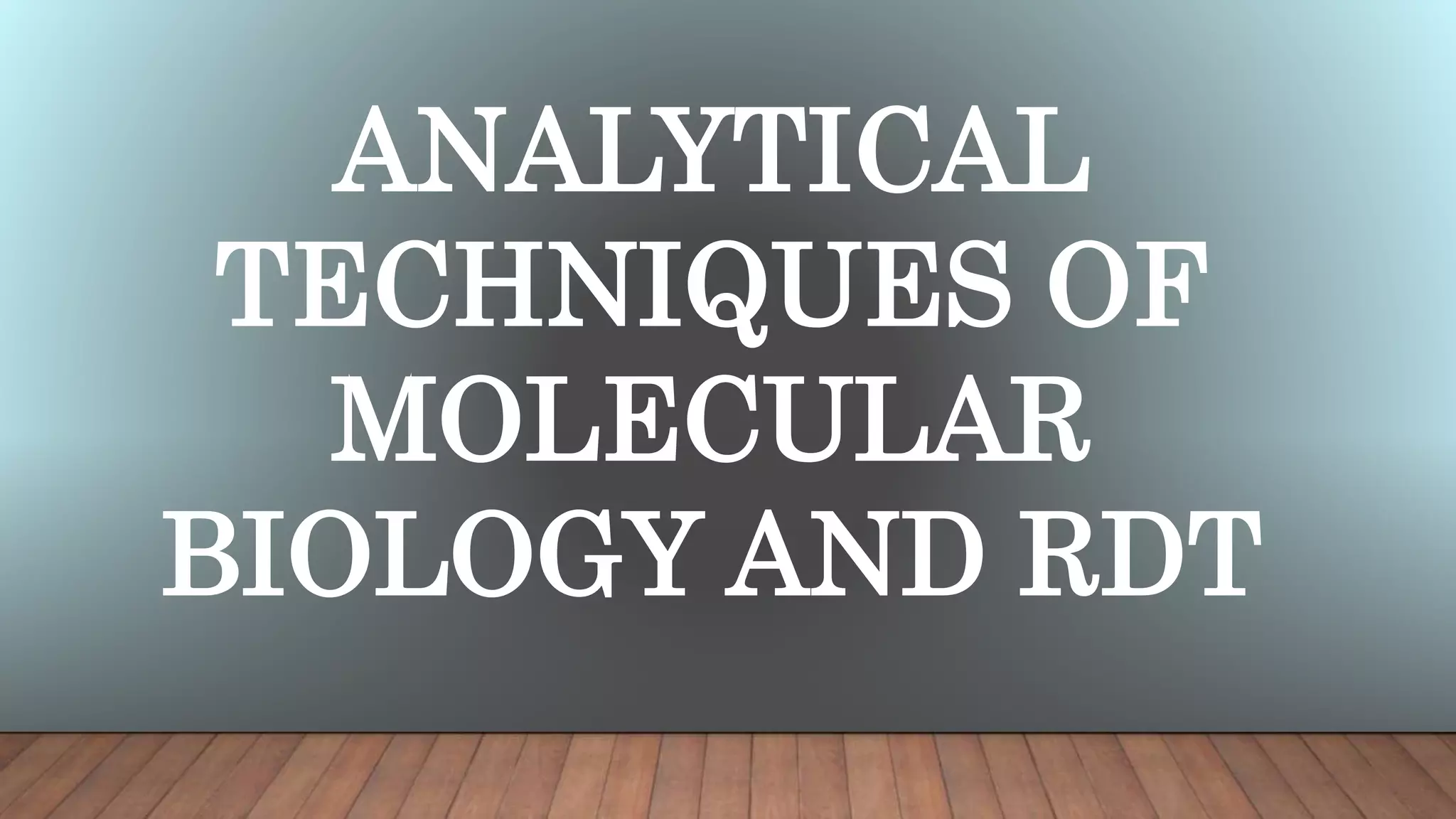
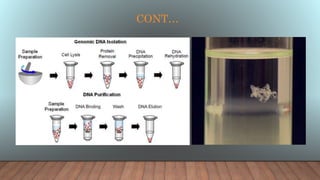
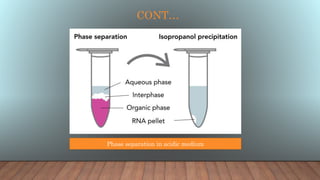
The document outlines various analytical techniques for DNA and RNA isolation and purification, including methods like organic extraction and the trizol method. Key steps involve cell lysis, removal of proteins and debris, and RNA degradation prevention, emphasizing the need for an RNase-free environment. It also discusses the quality assessment of nucleic acids using spectrophotometry and gel electrophoresis.