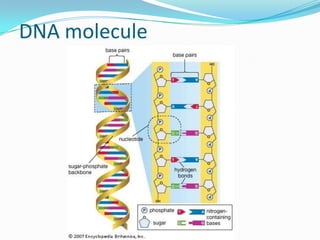
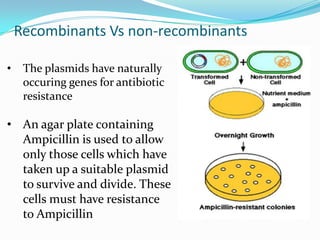
The document introduces DNA technology techniques, including DNA extraction, cloning, PCR, and fingerprinting, while explaining their applications. It covers the structure of nucleic acids, the process of gene cloning using vectors and hosts, PCR's role in DNA amplification, and methods of DNA profiling for forensics and paternity testing. Additionally, it discusses the use of PCR for pathogen detection in medical diagnoses.