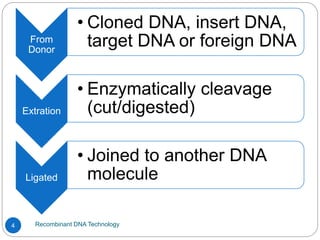
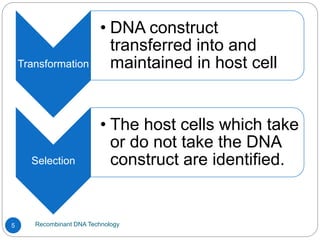
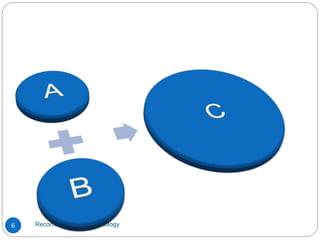
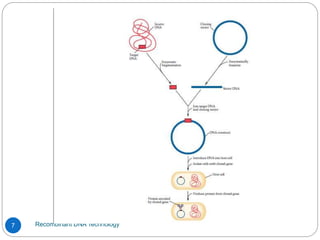
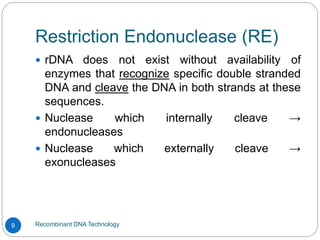
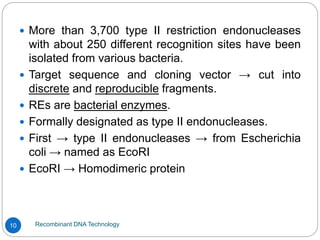
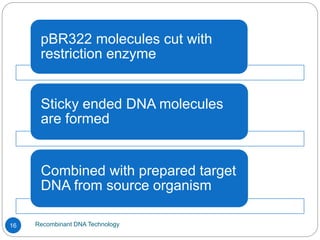
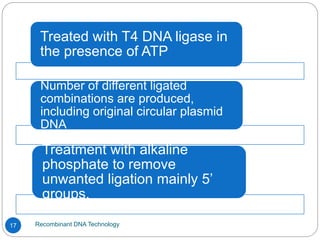
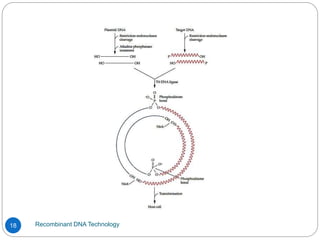
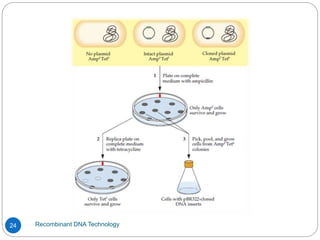
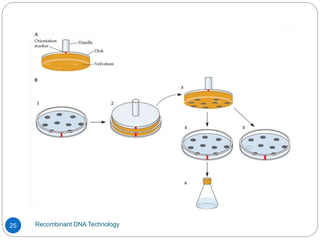
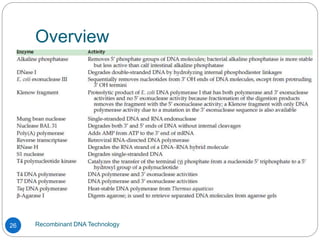
Recombinant DNA (rDNA) technology, also known as molecular cloning, involves combining DNA from different species to create new genetic combinations beneficial for various fields. The process includes enzymatic cleavage, transformation into host cells, and selection of transformed cells. Historical milestones highlight key developments in the technology, including the discovery of DNA structure and the creation of plasmids as cloning vectors.