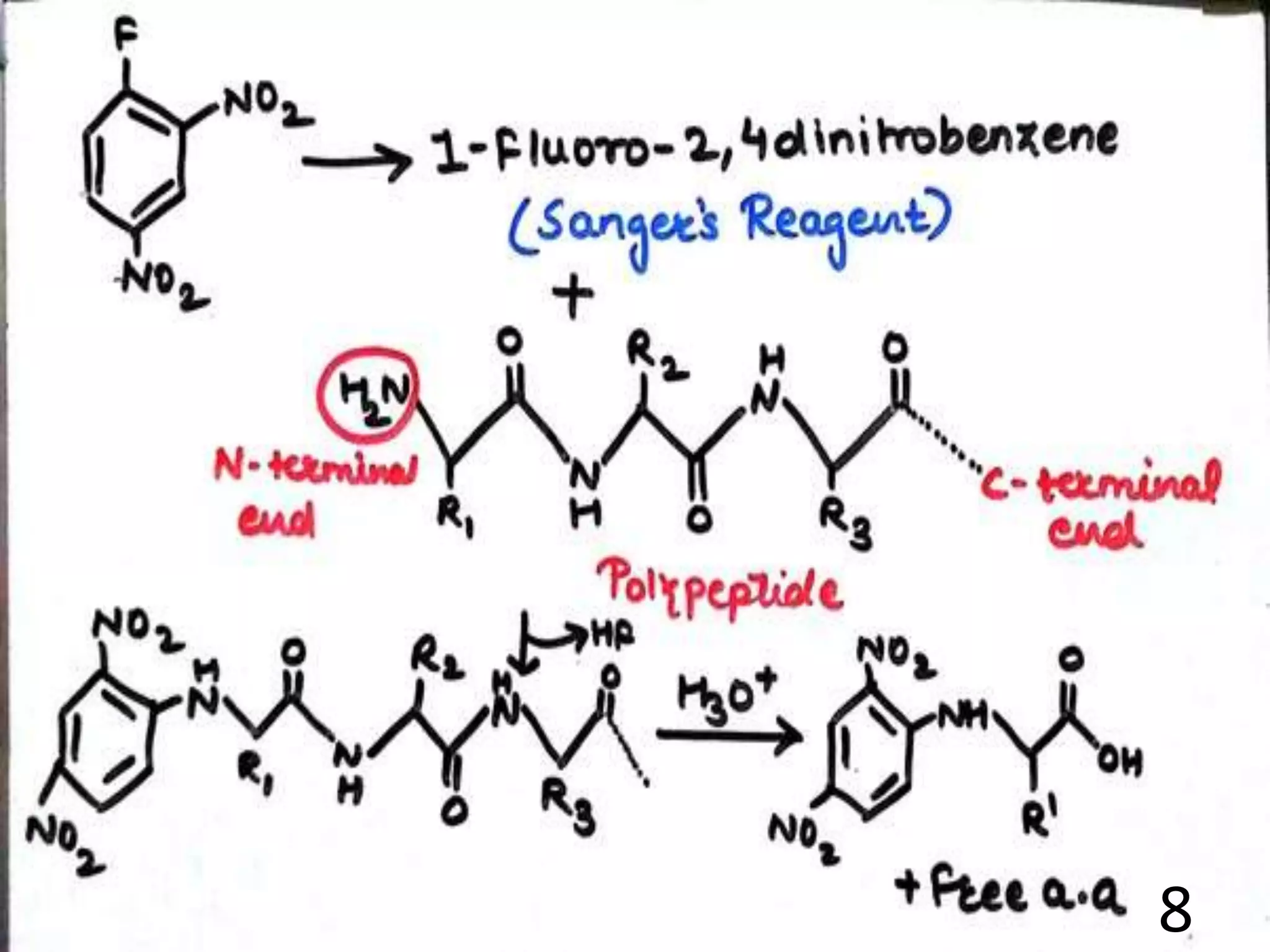
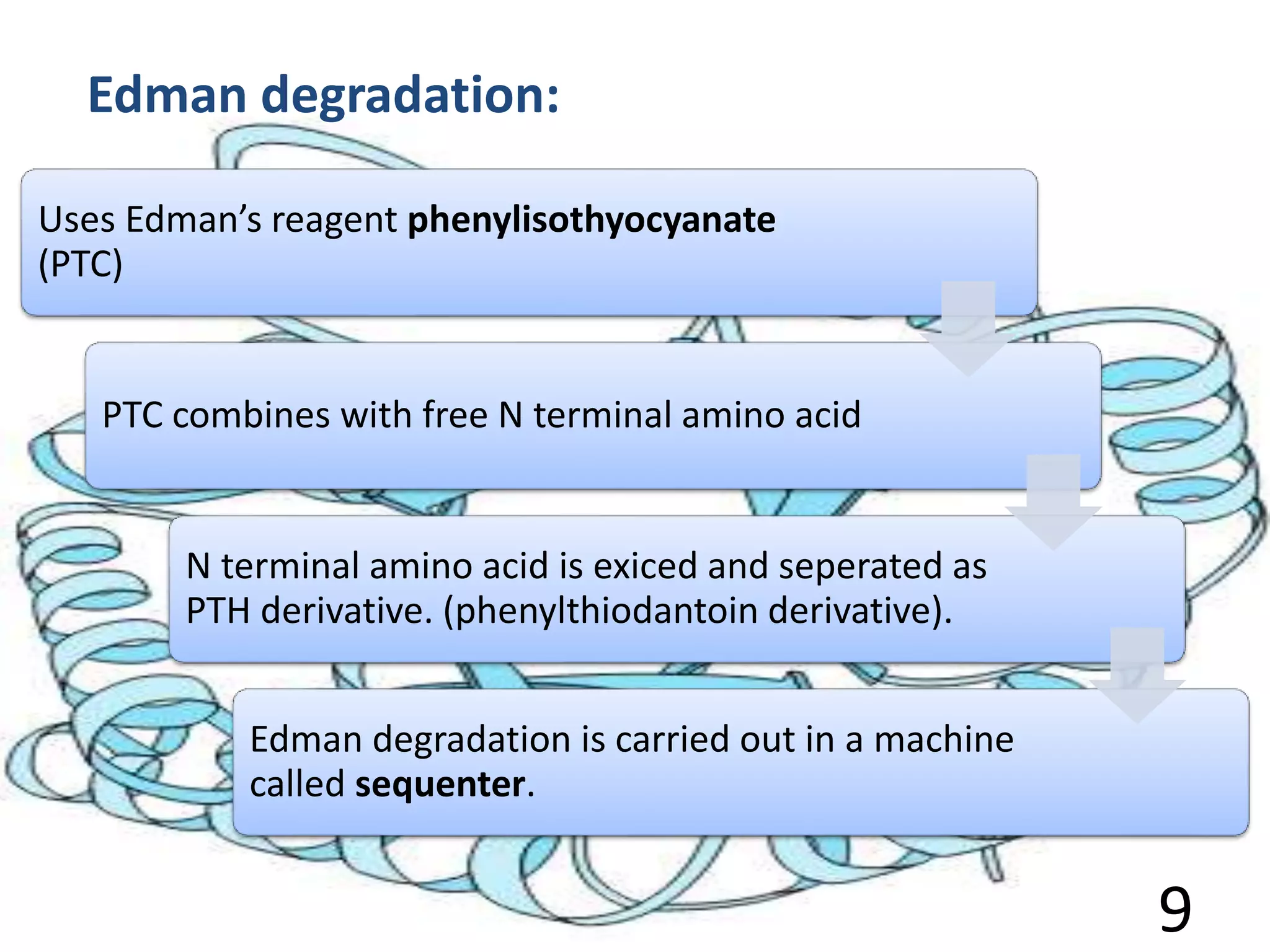
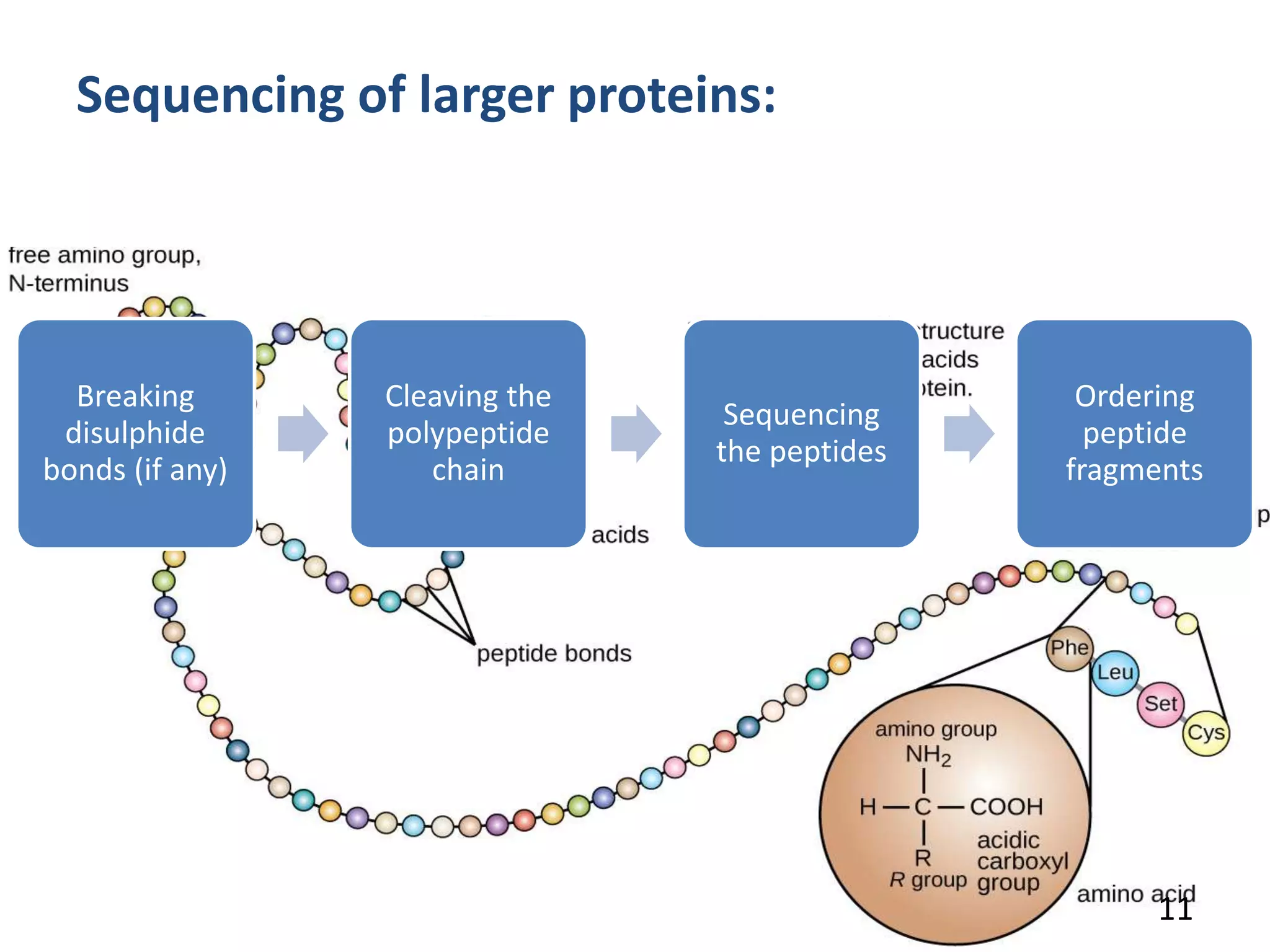
Fredric Sanger determined the first protein sequence in 1953, which was insulin. There are two main methods for protein sequencing - N-terminal sequencing including the Sanger method and Edman degradation, and C-terminal sequencing using carboxypeptidases. Protein sequences can be predicted from DNA sequences after determining the gene sequence. Comparing protein sequences between organisms can provide information about evolutionary relationships and how organisms have diverged over time as mutations cause amino acid changes. Work has been done in Pakistan on sequencing various proteins.