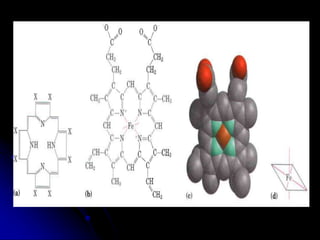
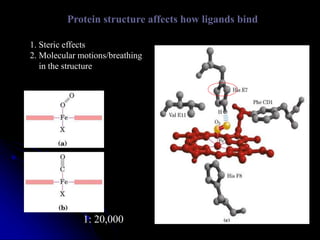
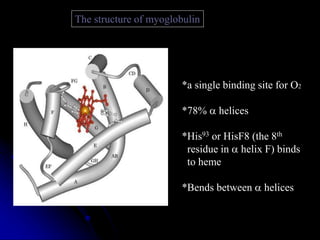
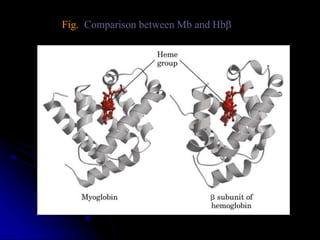
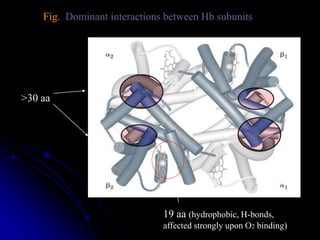
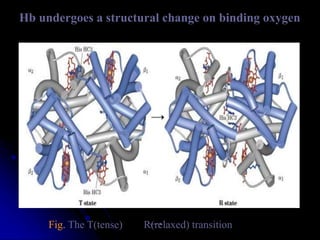
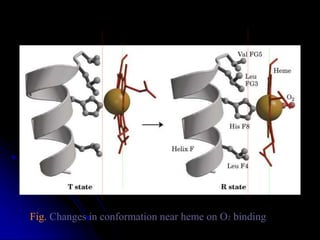
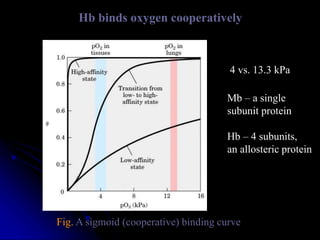
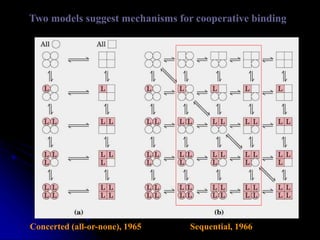
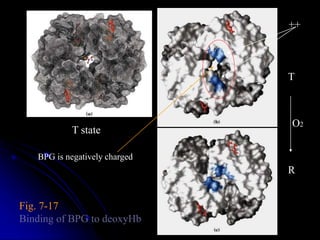
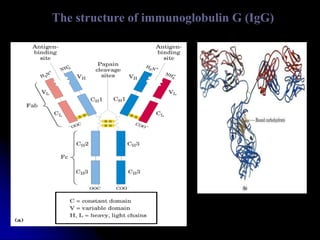
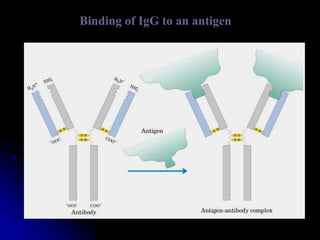
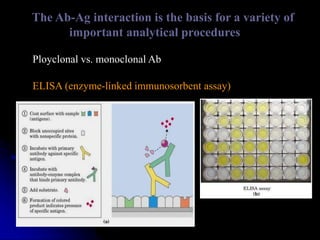
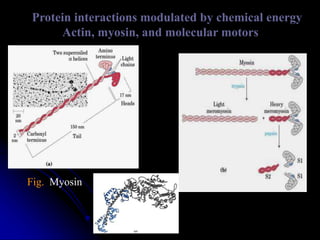
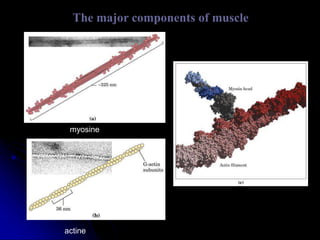
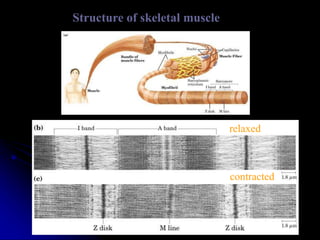
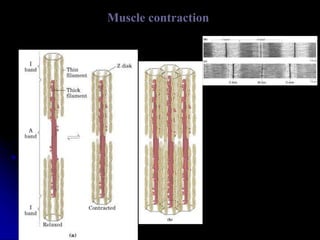
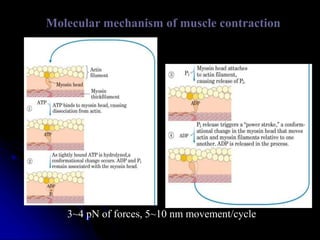
The document discusses protein-ligand and protein-protein interactions, highlighting their essential roles in biological functions and molecular processes like DNA replication. It explains oxygen binding in hemoglobin and myoglobin, detailing their structural characteristics and cooperative binding mechanisms. Additionally, it covers the structure of immunoglobulin G in antigen interaction and the mechanics of muscle contraction involving actin and myosin.