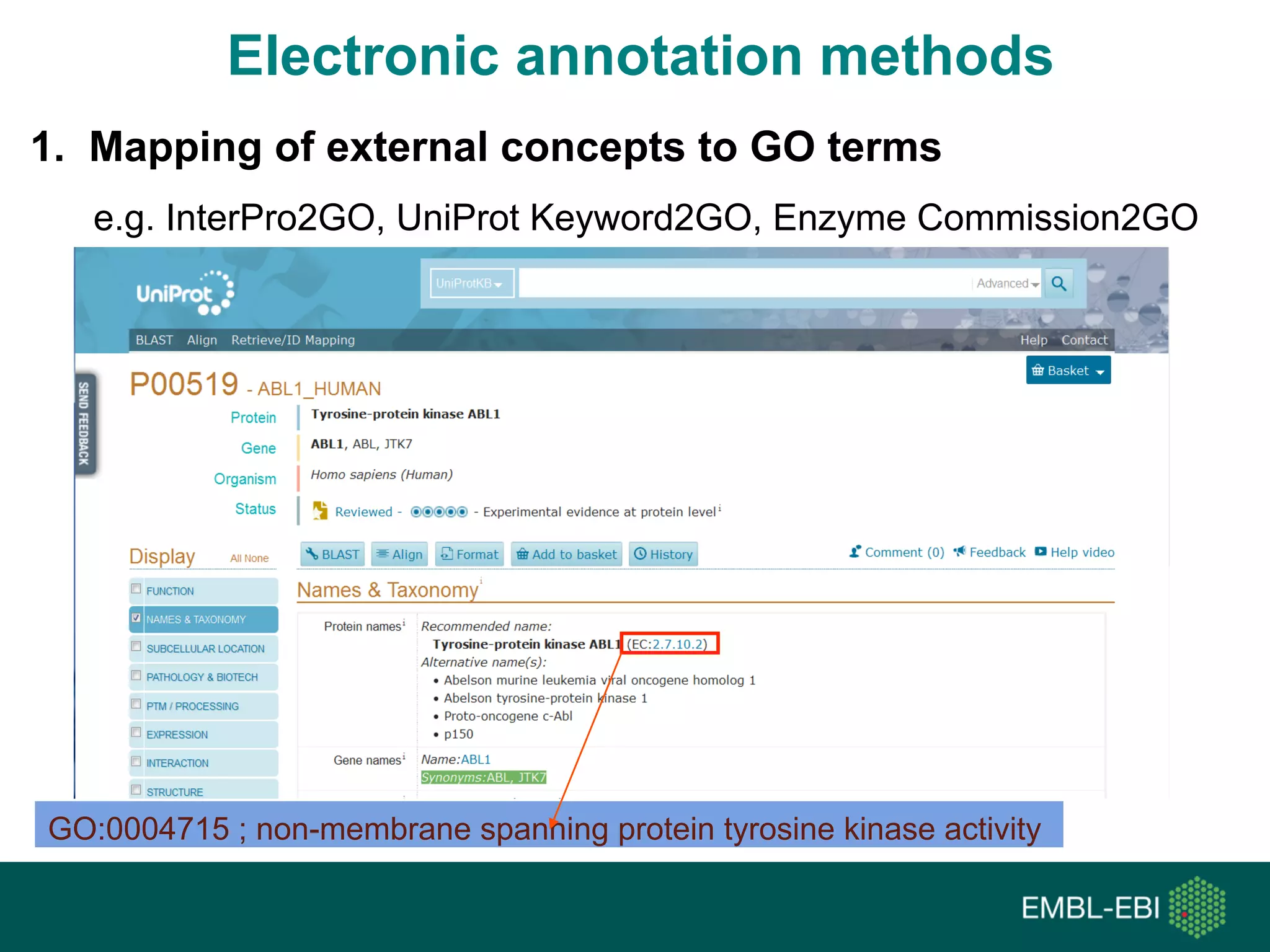
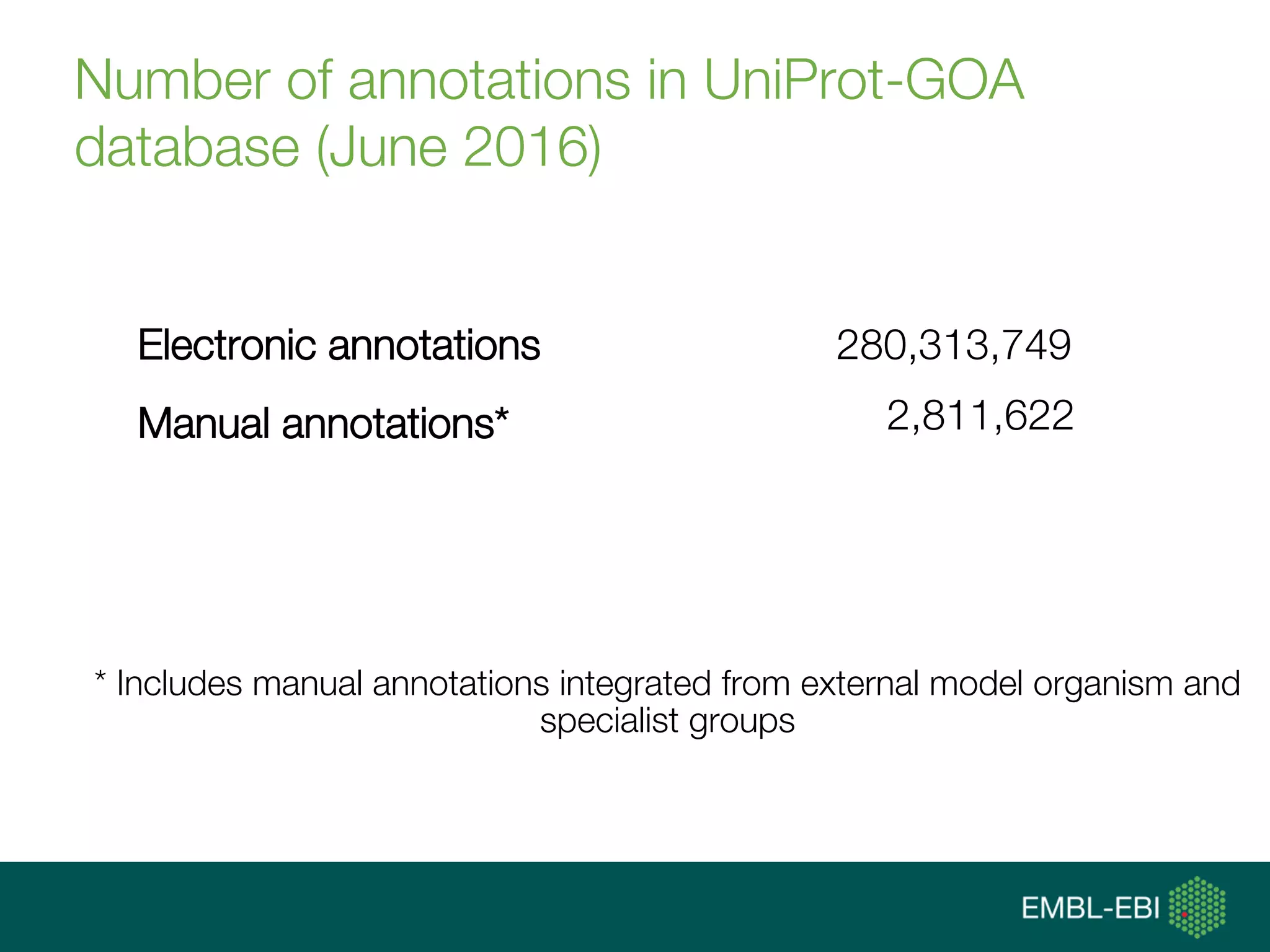
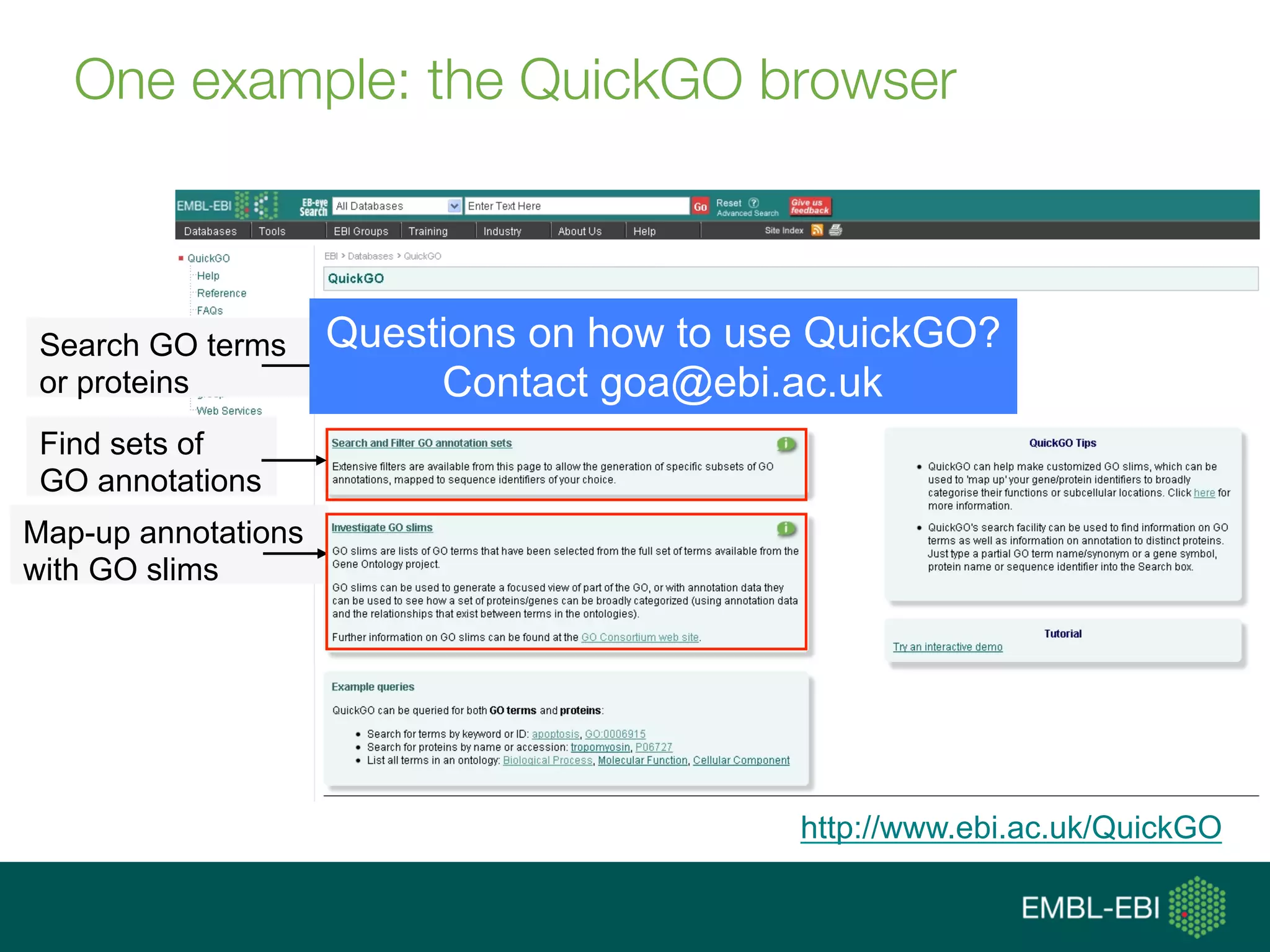
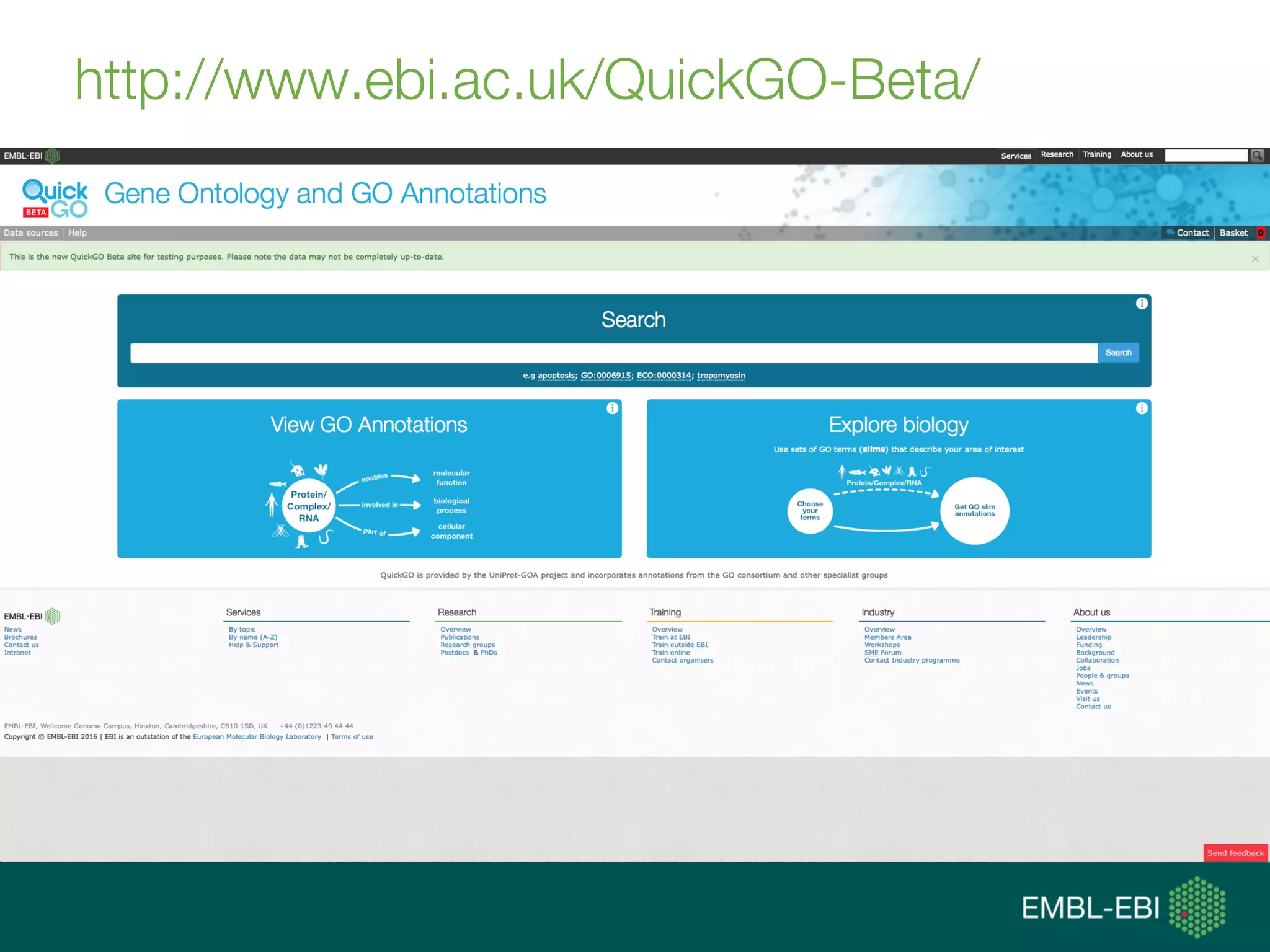
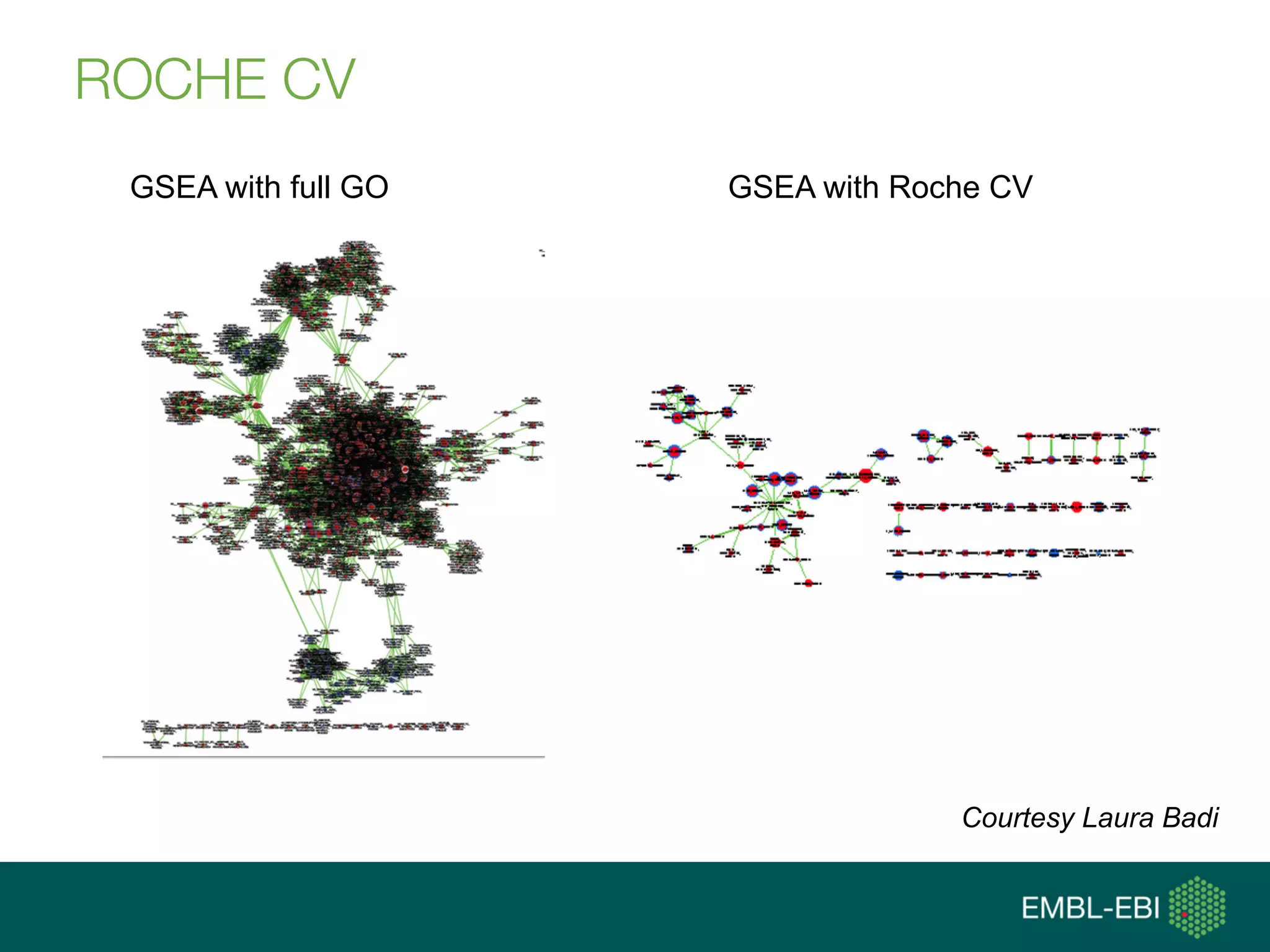
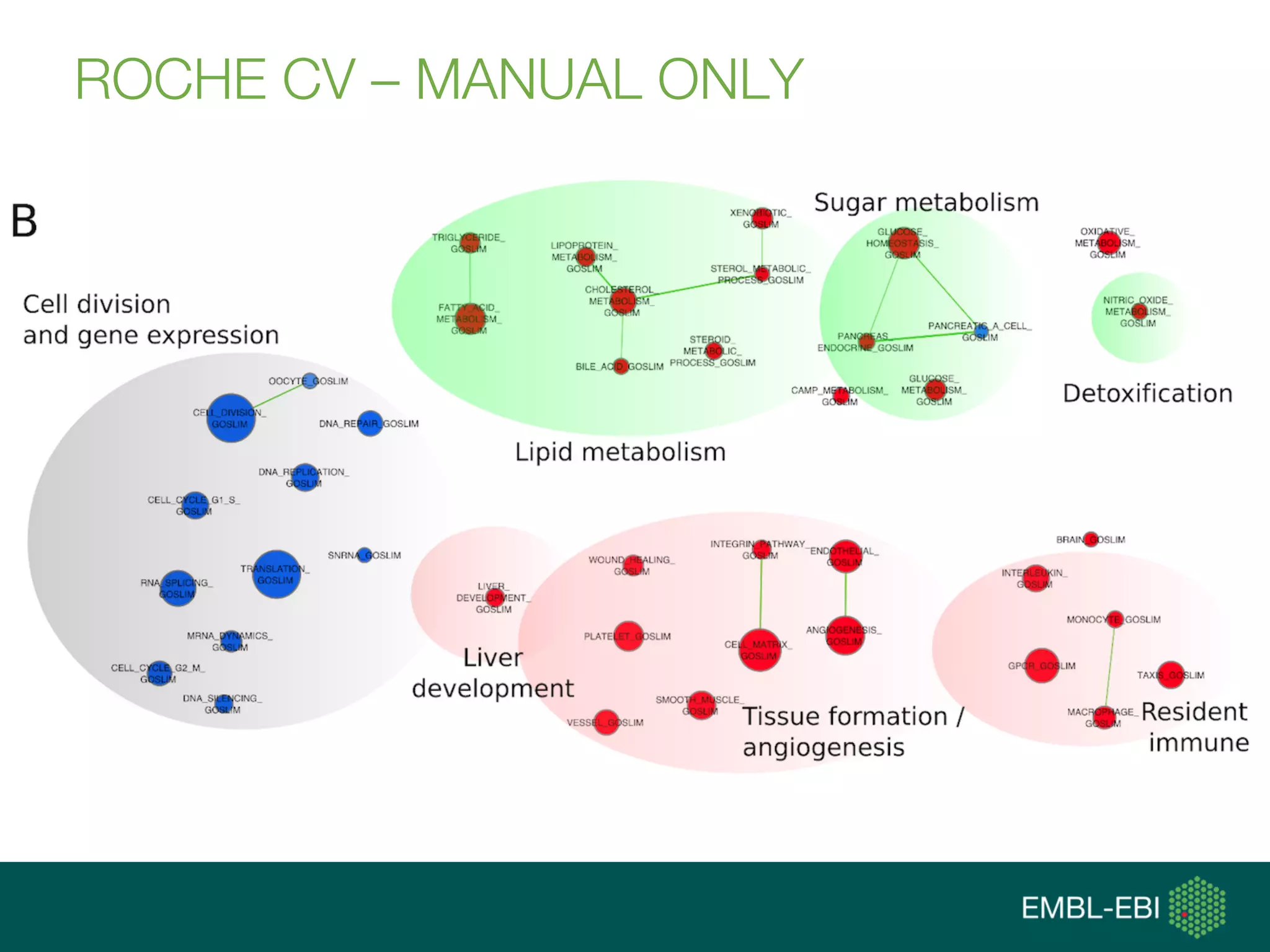
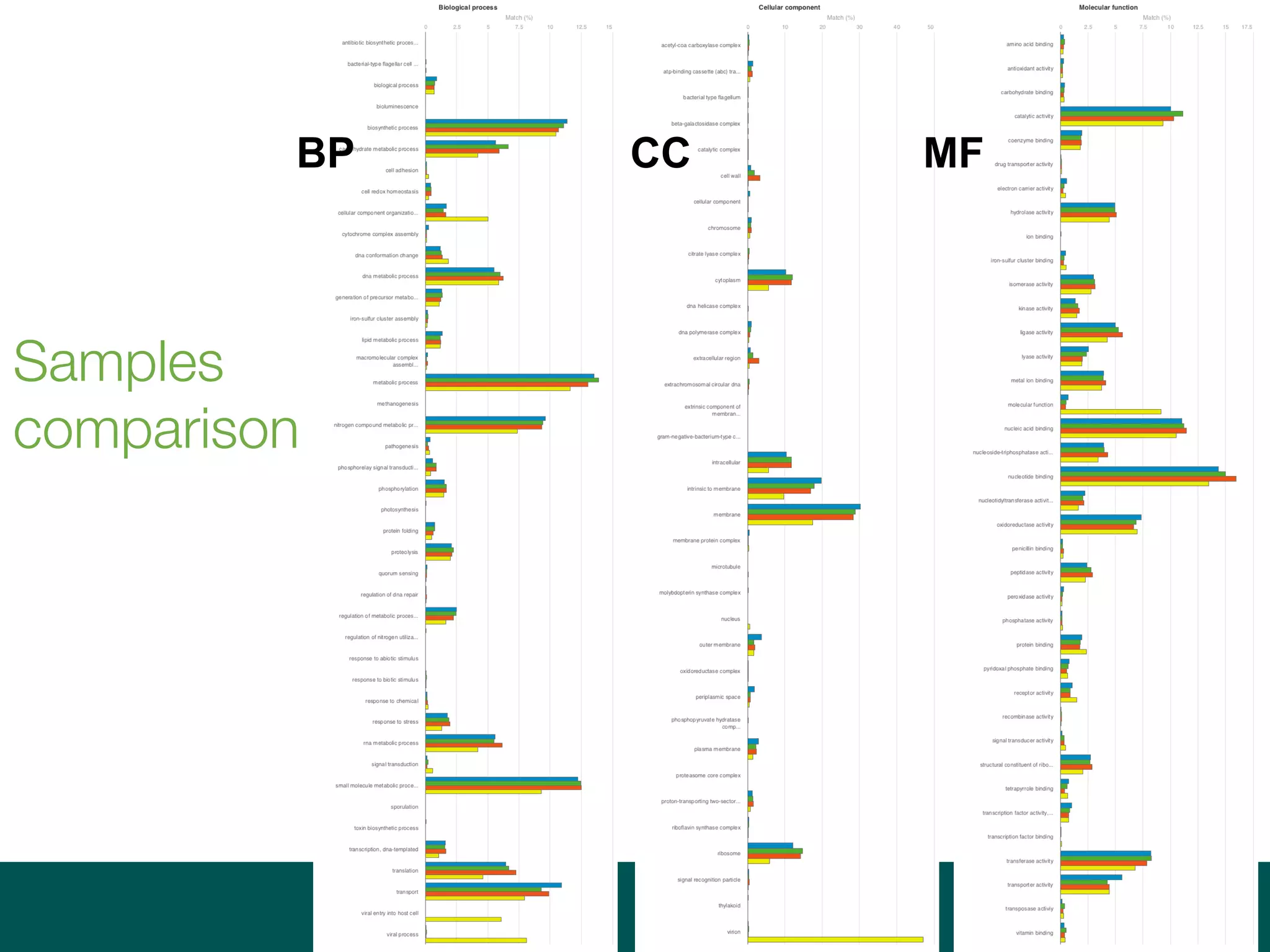
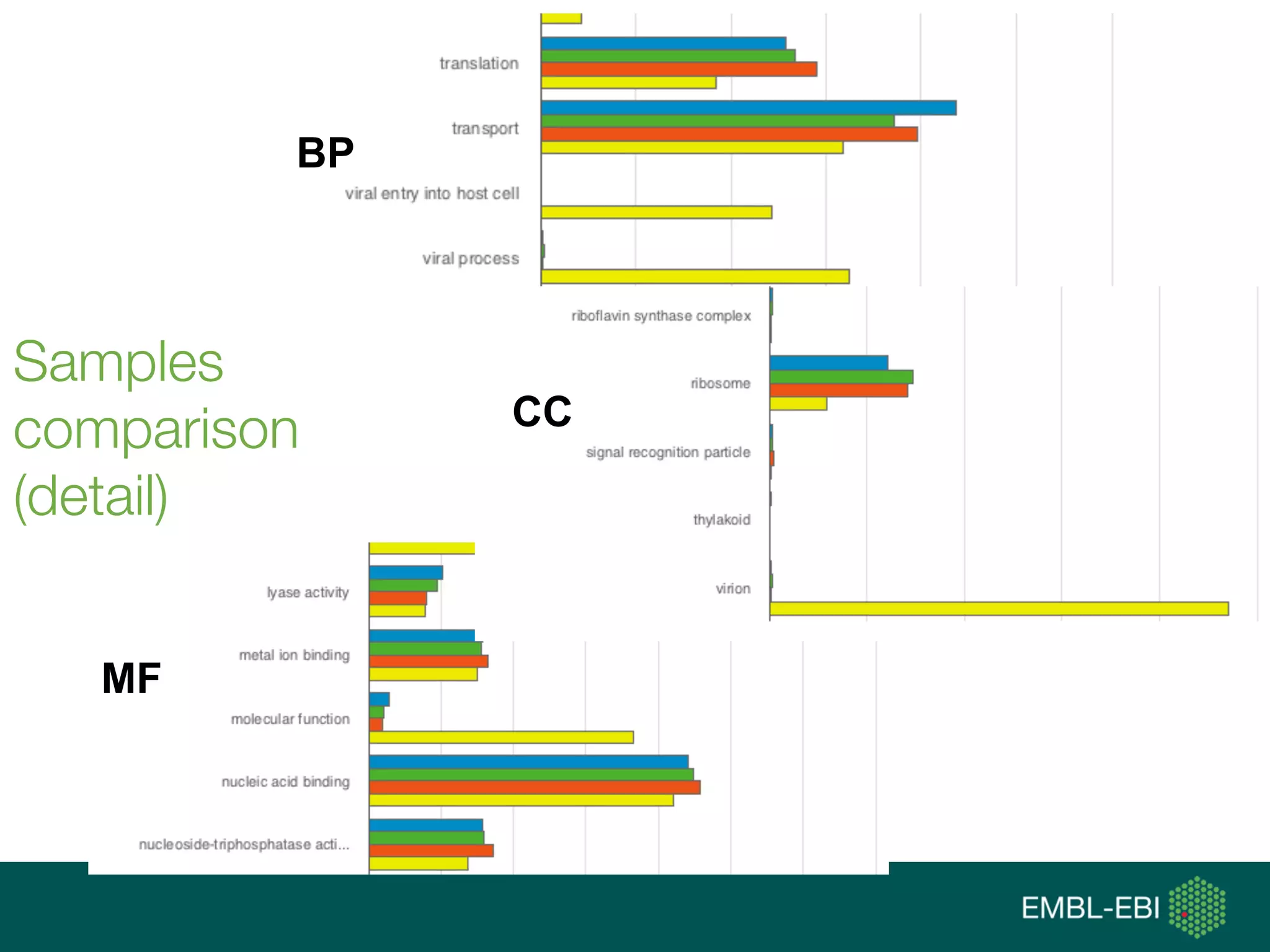
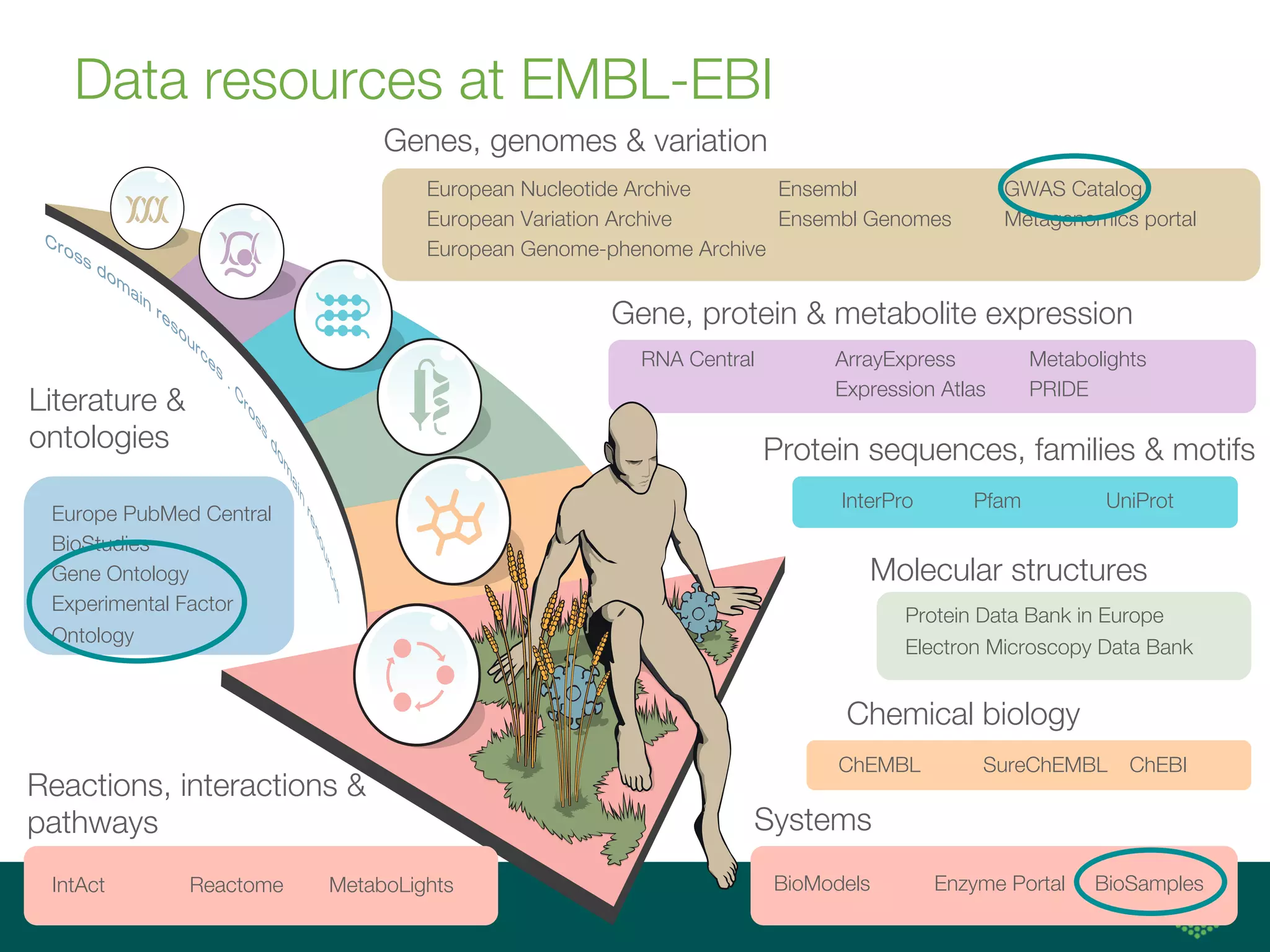
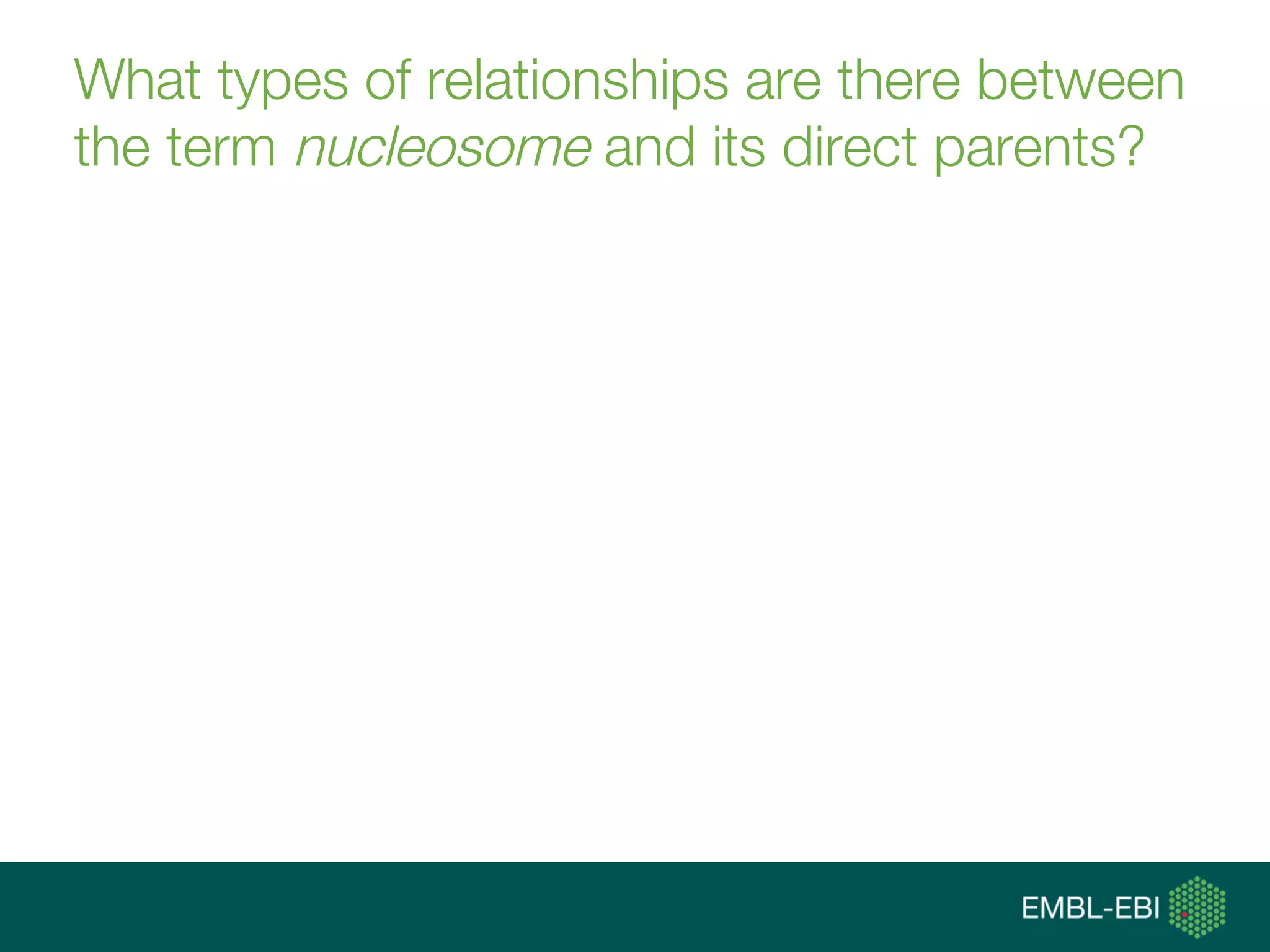
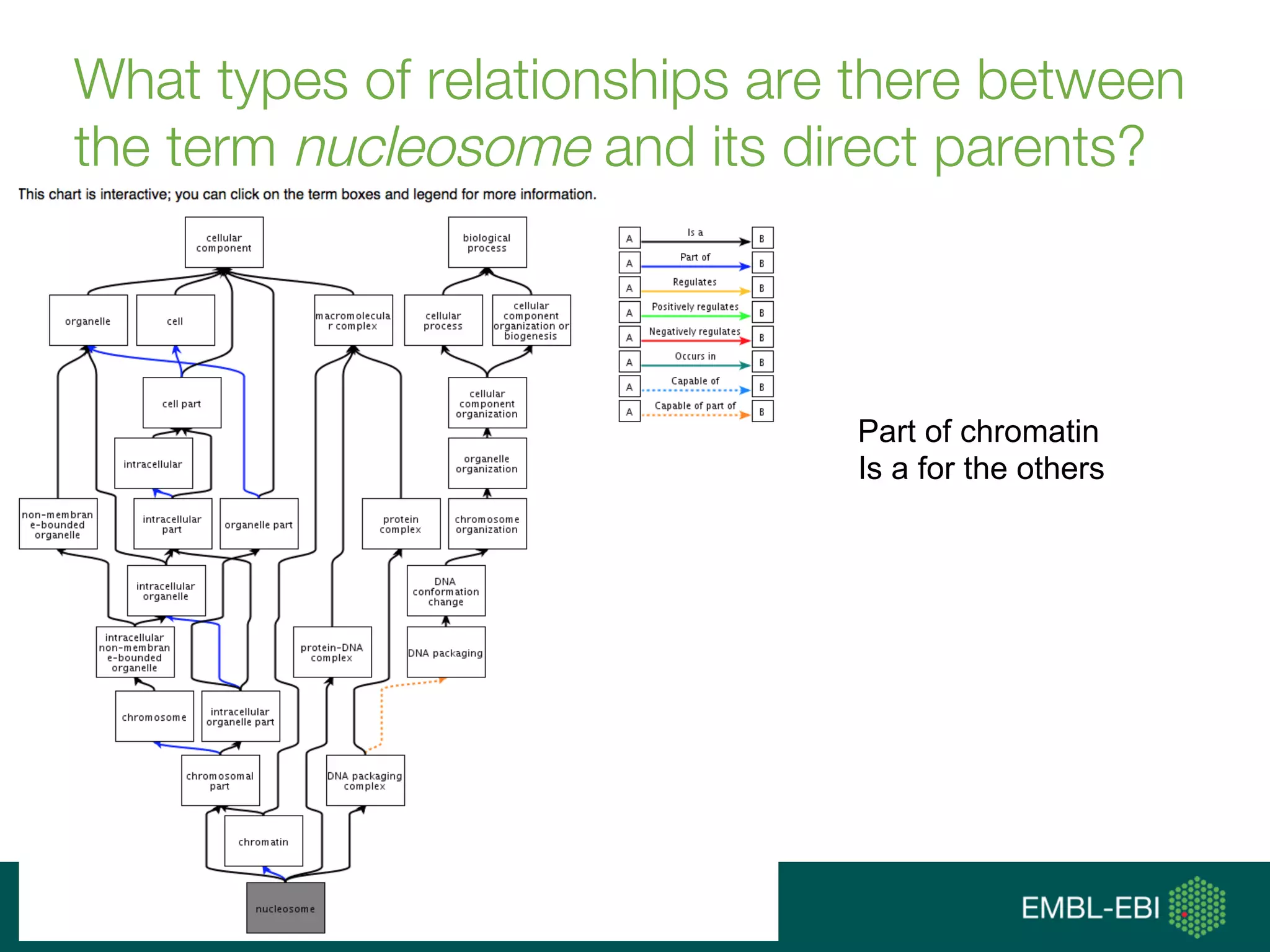
Ontologies for life sciences: examples from the Gene Ontology
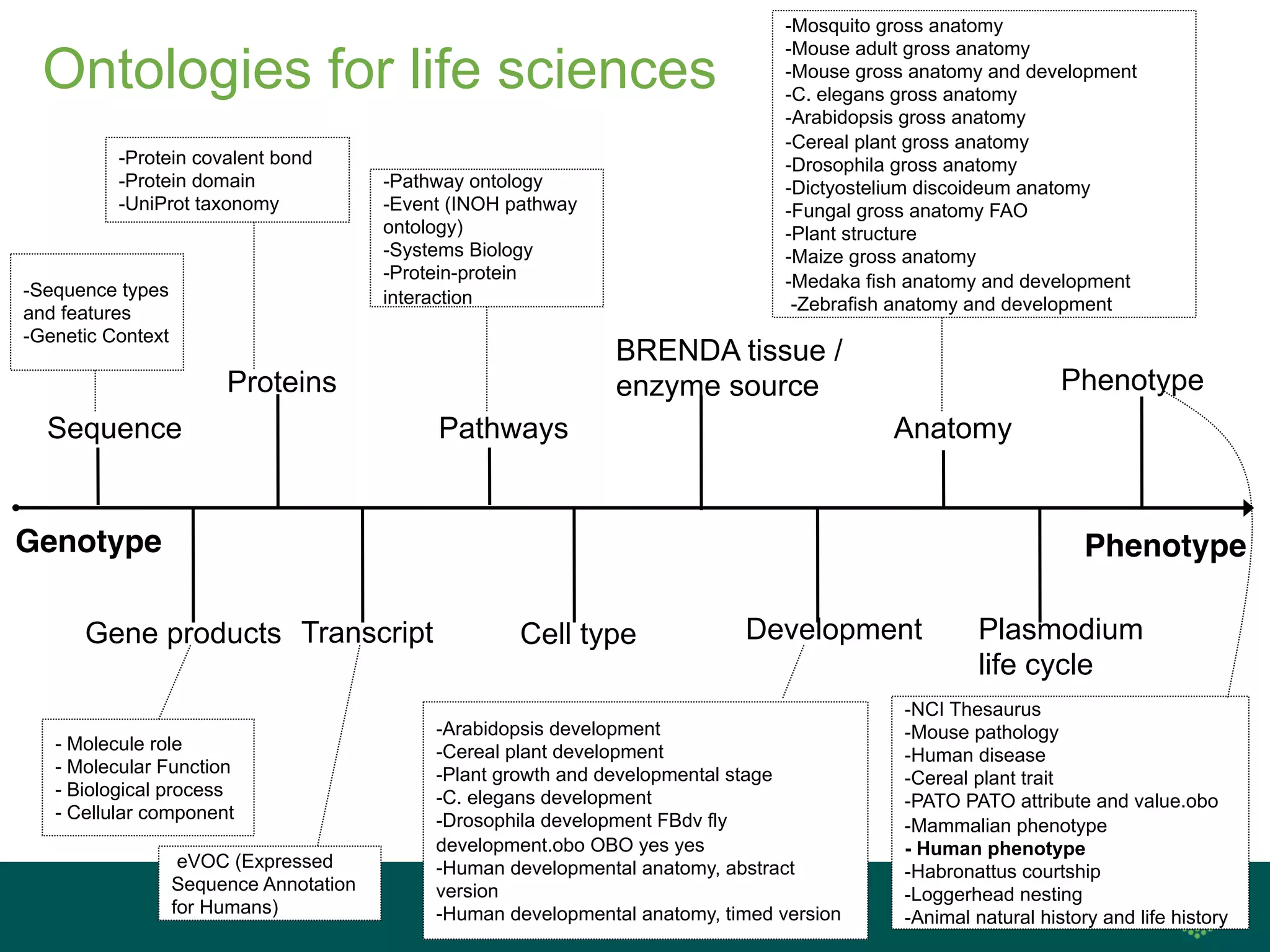
The document discusses ontologies for life sciences, using the Gene Ontology (GO) as an example. It provides an overview of GO, describing it as a way to capture biological knowledge for gene products in a written and computable form using a set of concepts and relationships arranged hierarchically. GO allows consistent descriptions of genes/gene products across databases. Model organism databases provide annotations connecting genes to GO terms. The GO is a collaborative effort to address the need for consistent descriptions of genes.


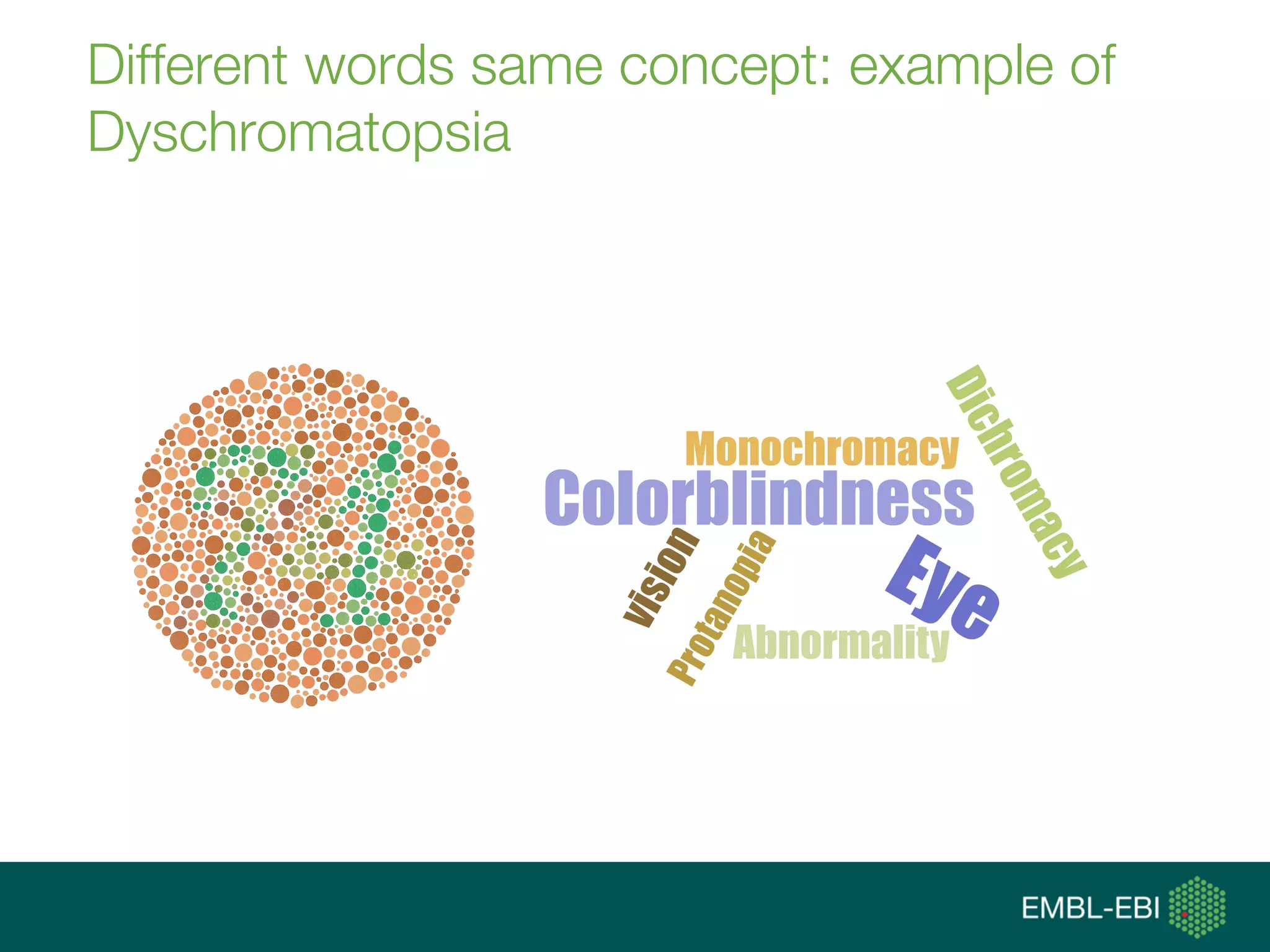

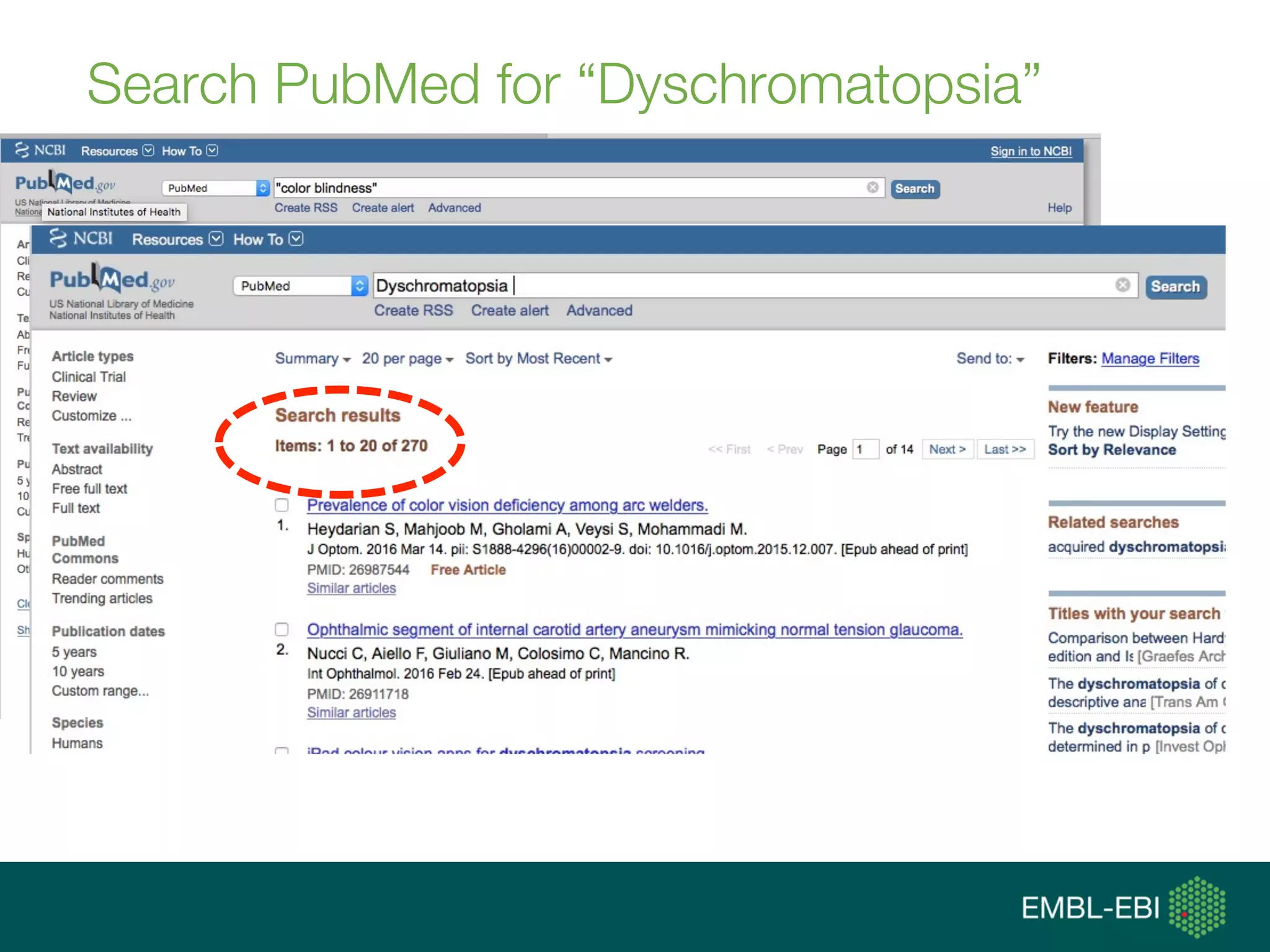



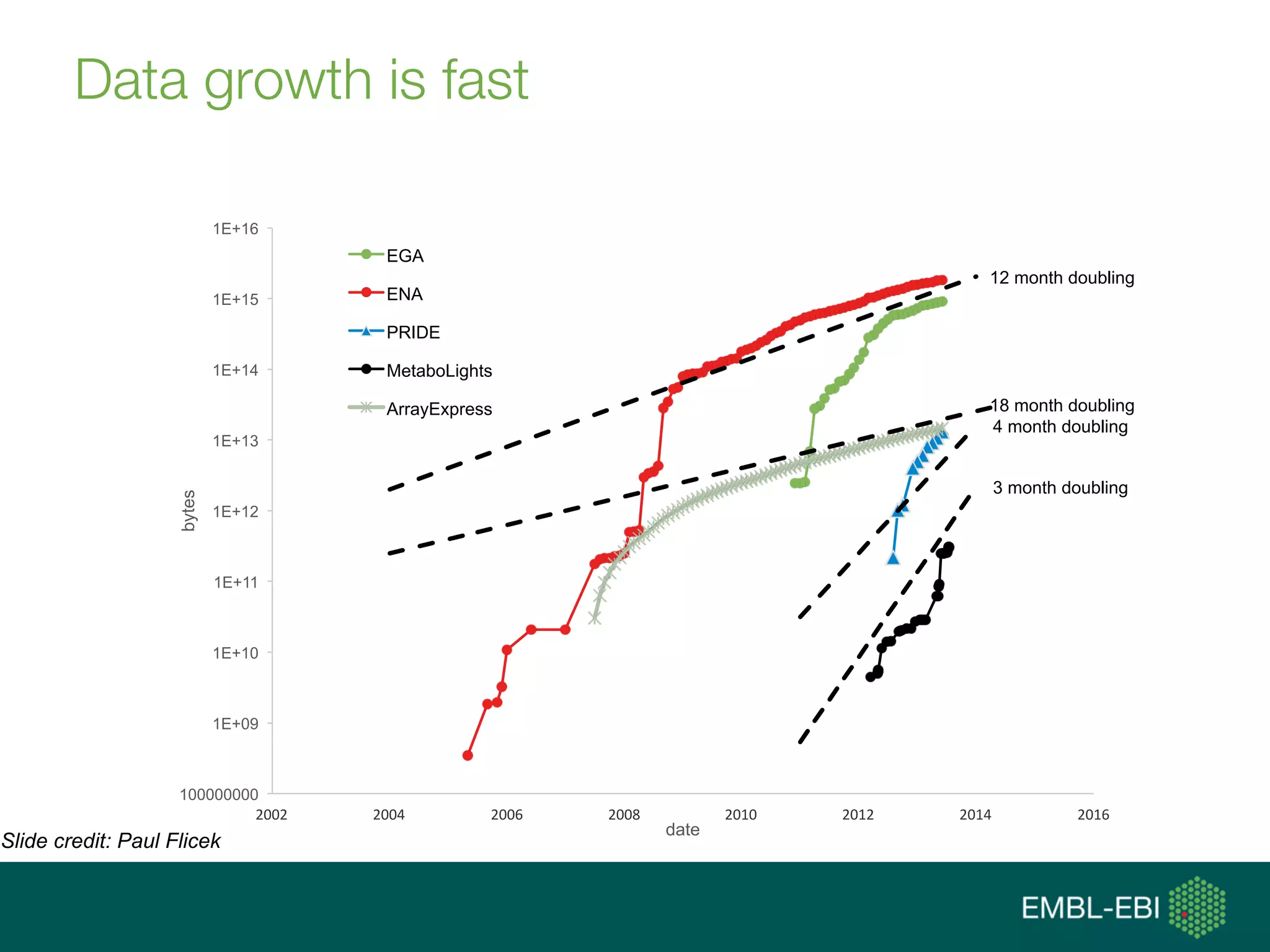

![data integration [ˈdeɪtə ˌɪntəˈgreɪʃən]:
(computational) means to access, retrieve
and analyse data sets from different
sources in order to exploit them, i.e., gain
new knowledge, and share that new
knowledge](https://image.slidesharecdn.com/tgaccourtot-160919073248/75/Ontologies-for-life-sciences-examples-from-the-gene-ontology-13-2048.jpg)
![data integration [ˈdeɪtə ˌɪntəˈgreɪʃən]:
(computational) means to access, retrieve
and analyse data sets from different
sources in order to exploit them, i.e., gain
new knowledge, and share that new
knowledge](https://image.slidesharecdn.com/tgaccourtot-160919073248/75/Ontologies-for-life-sciences-examples-from-the-gene-ontology-14-2048.jpg)


























































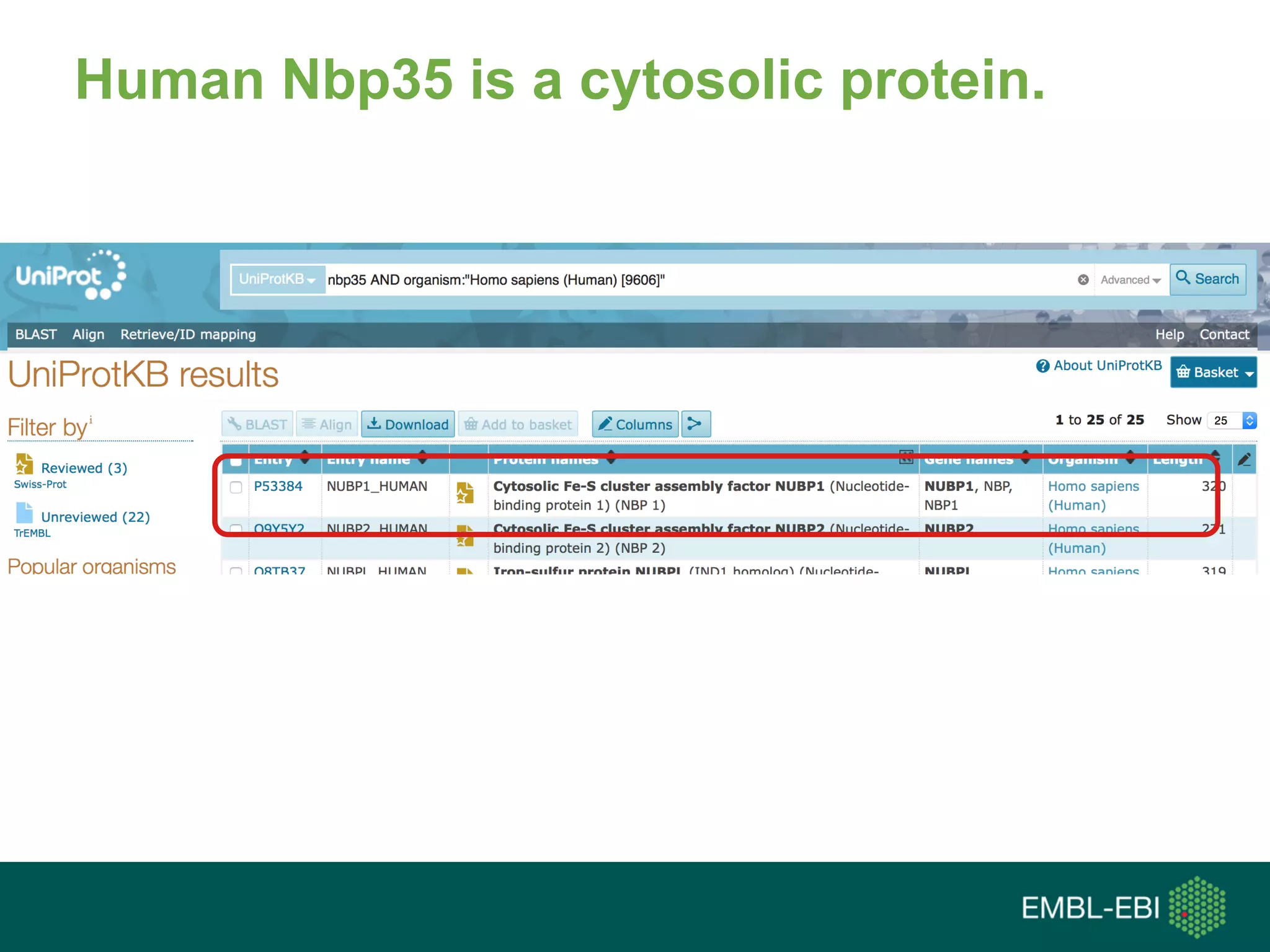
![FIG. 2. Human Nbp35 is a cytosolic
protein. (A) EGFP fluorescence of a HeLa
cell transiently transfected with a vector
encoding a huNbp35-EGFP fusion protein
(right) in comparison to the endogenous
autofluorescence (AFL) of control cells
(left).
(C) Sub-cellular localization of huNbp35 by cell fractionation. […]HuNbp35
exclusively colocalizes with tubulin in the cytosolic fraction, but not with
mitochondrial aconitase (mtAconitase) present in the membrane fraction.](https://image.slidesharecdn.com/tgaccourtot-160919073248/75/Ontologies-for-life-sciences-examples-from-the-gene-ontology-90-2048.jpg)






![GO evidence codes [small excerpt]
TAS, Traceable author statement
NAS, Non-traceable author statement
IDA, Inferred from Direct Assay
IMP, Inferred from Mutant Phenotype
IPI, Inferred from Physical Interaction
Experimental
evidence,
Methods &
Results
Abstract &
Introduction](https://image.slidesharecdn.com/tgaccourtot-160919073248/75/Ontologies-for-life-sciences-examples-from-the-gene-ontology-99-2048.jpg)