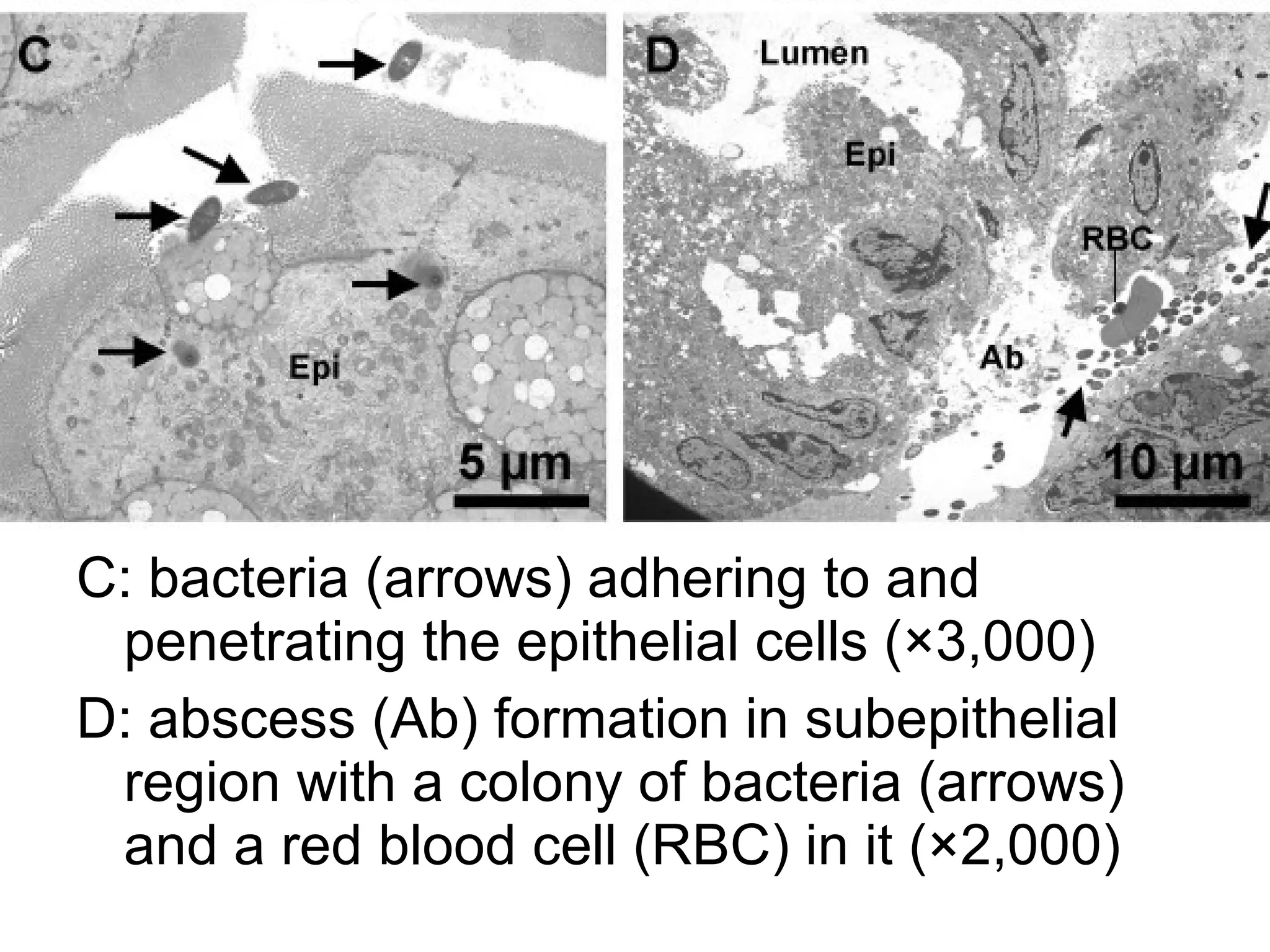
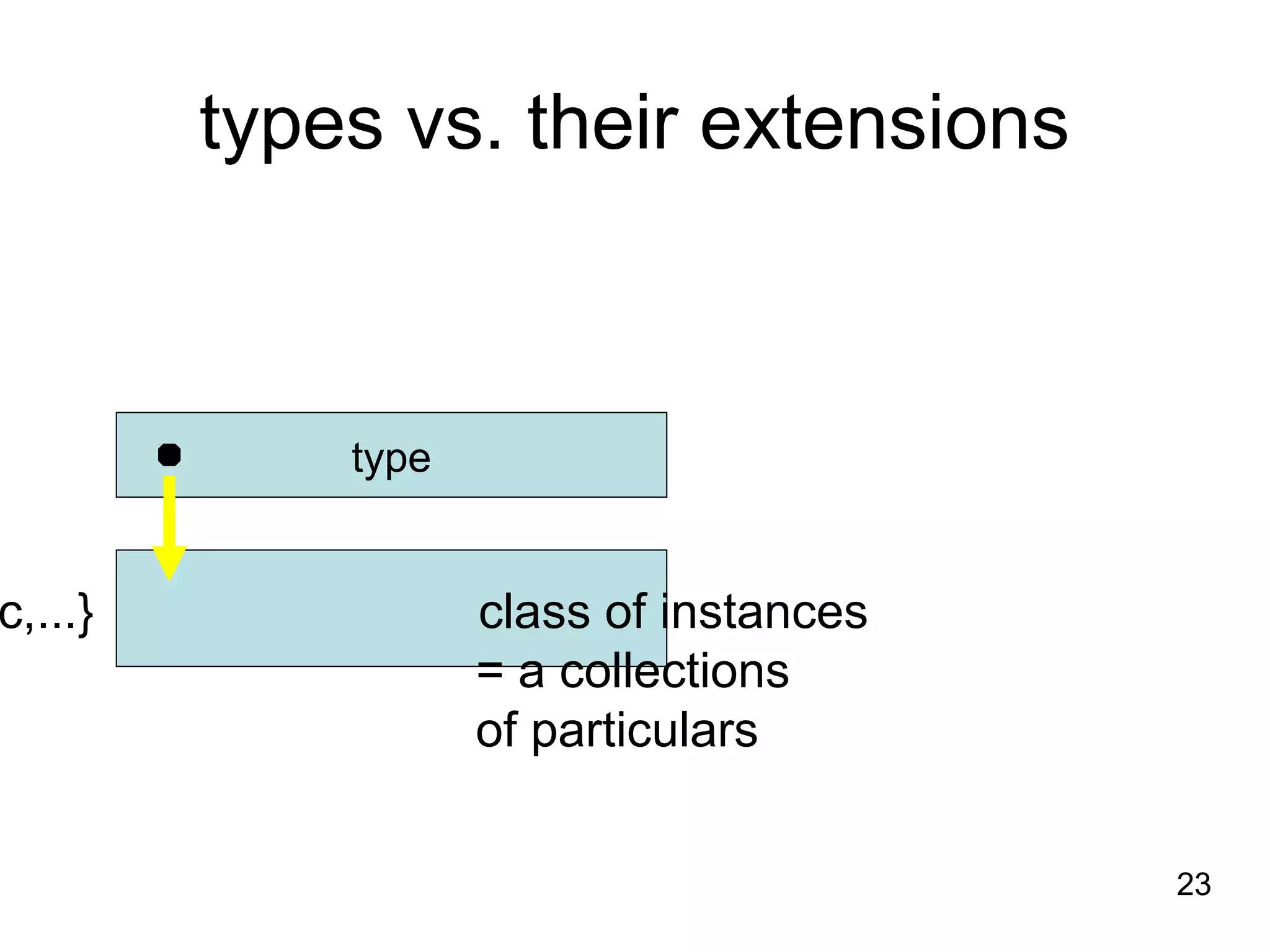
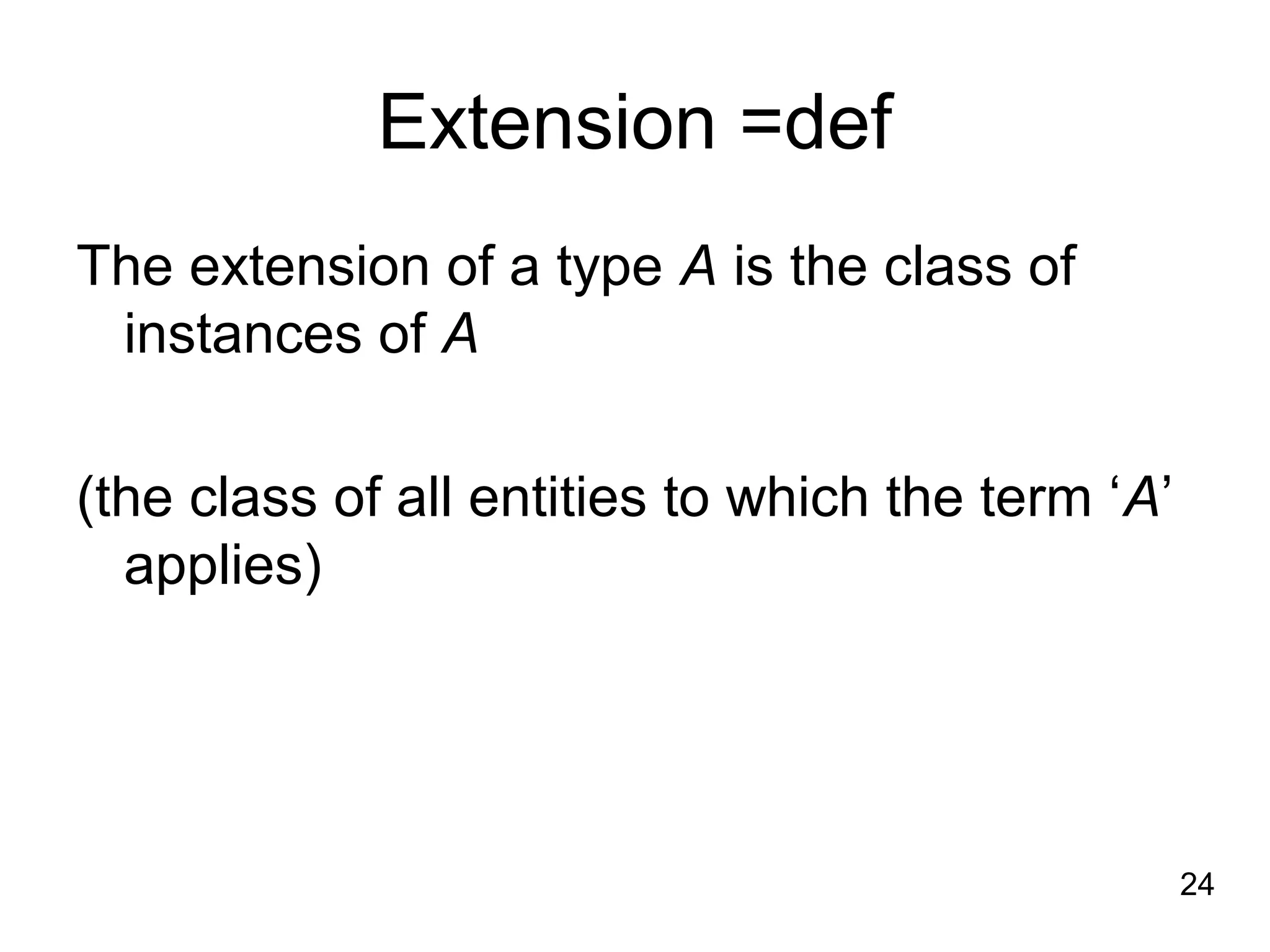
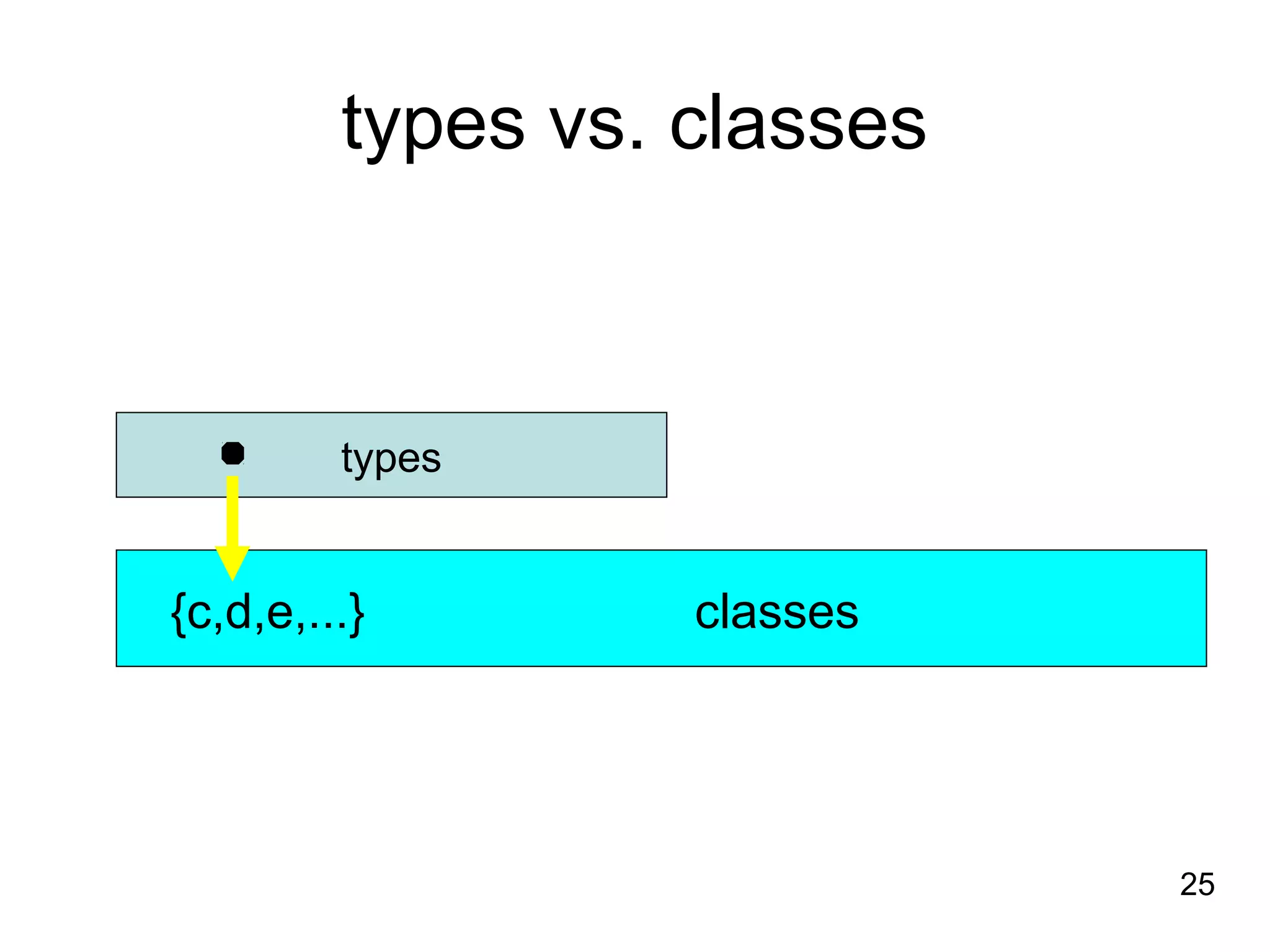
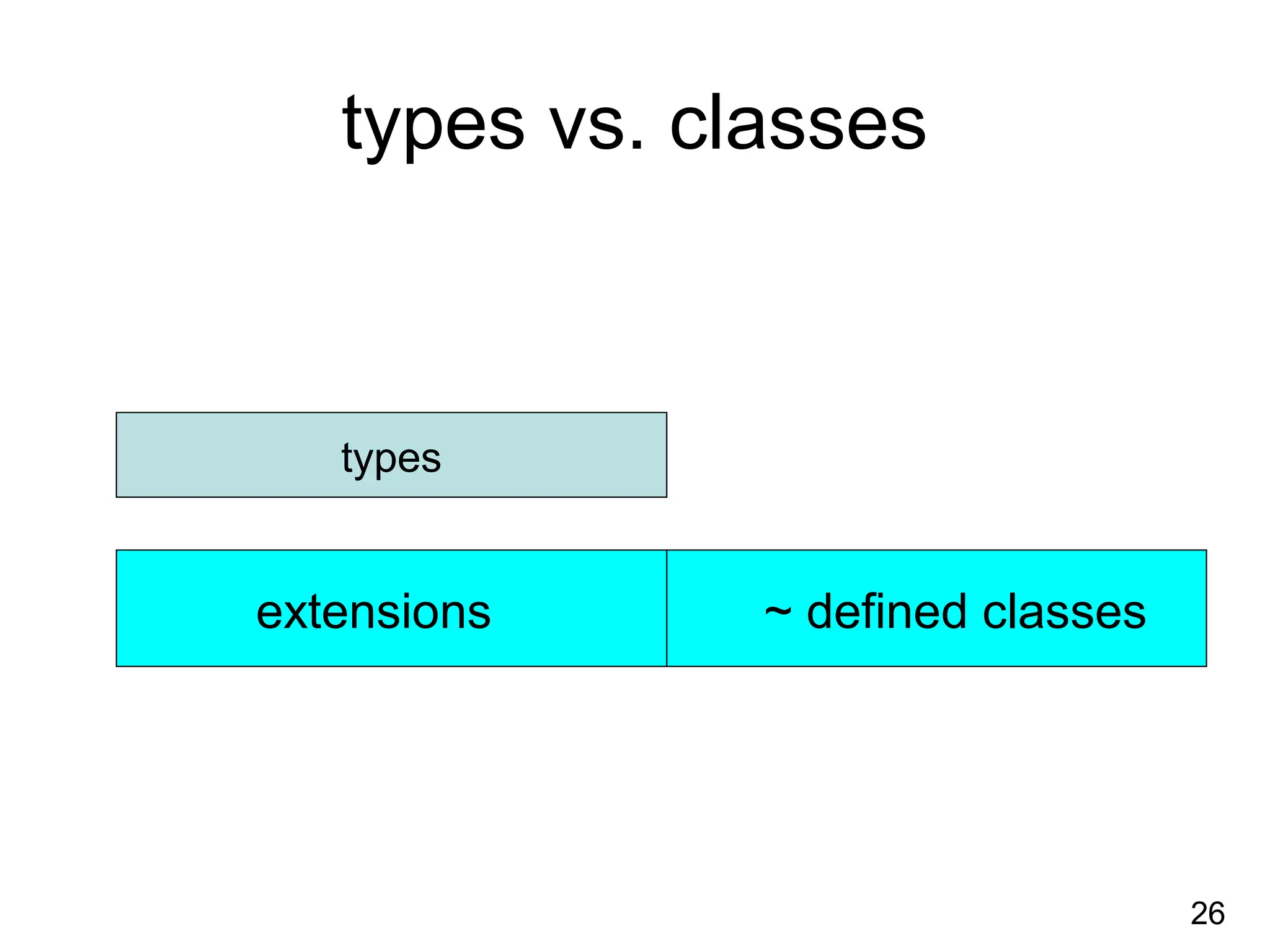
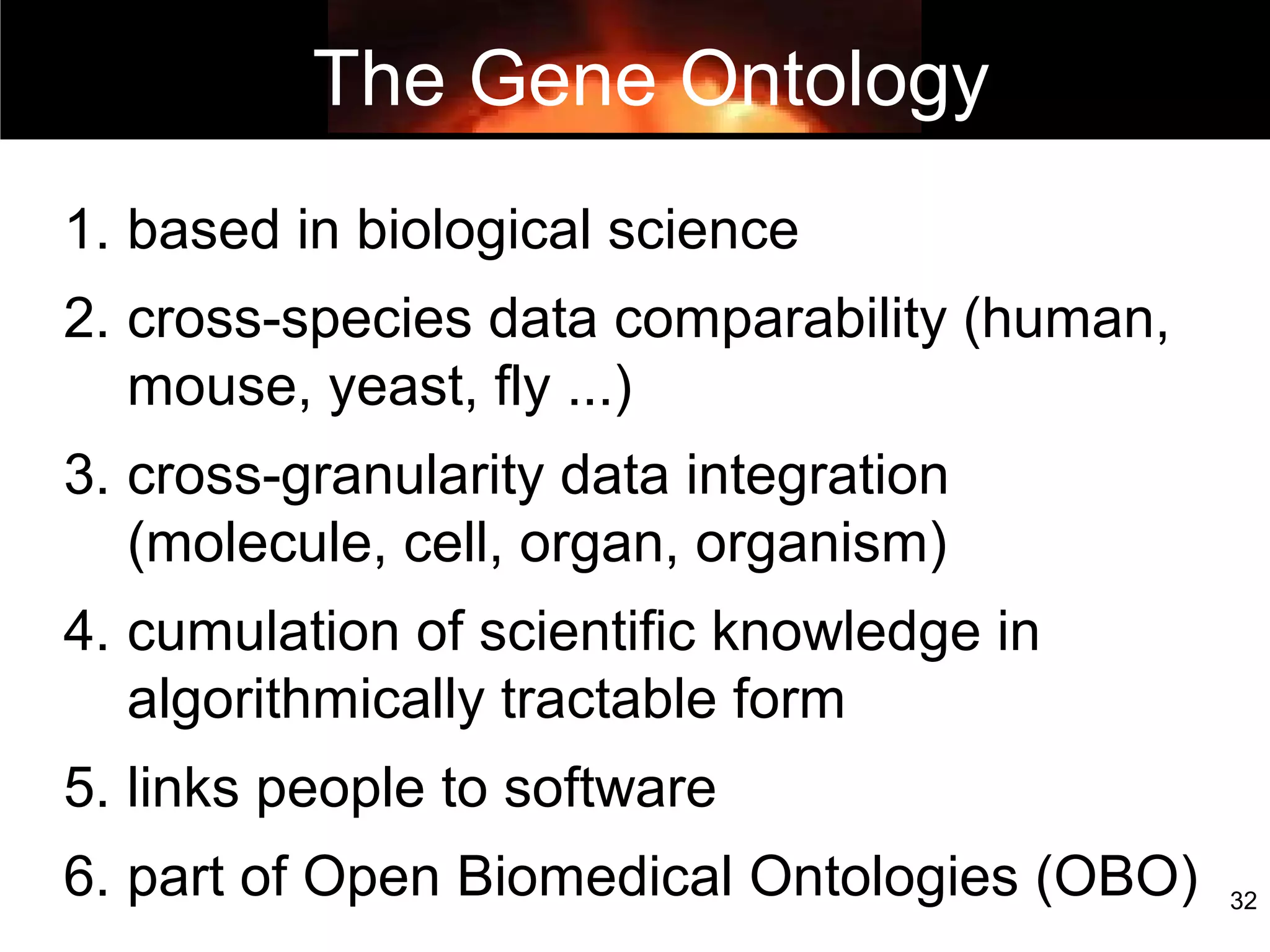
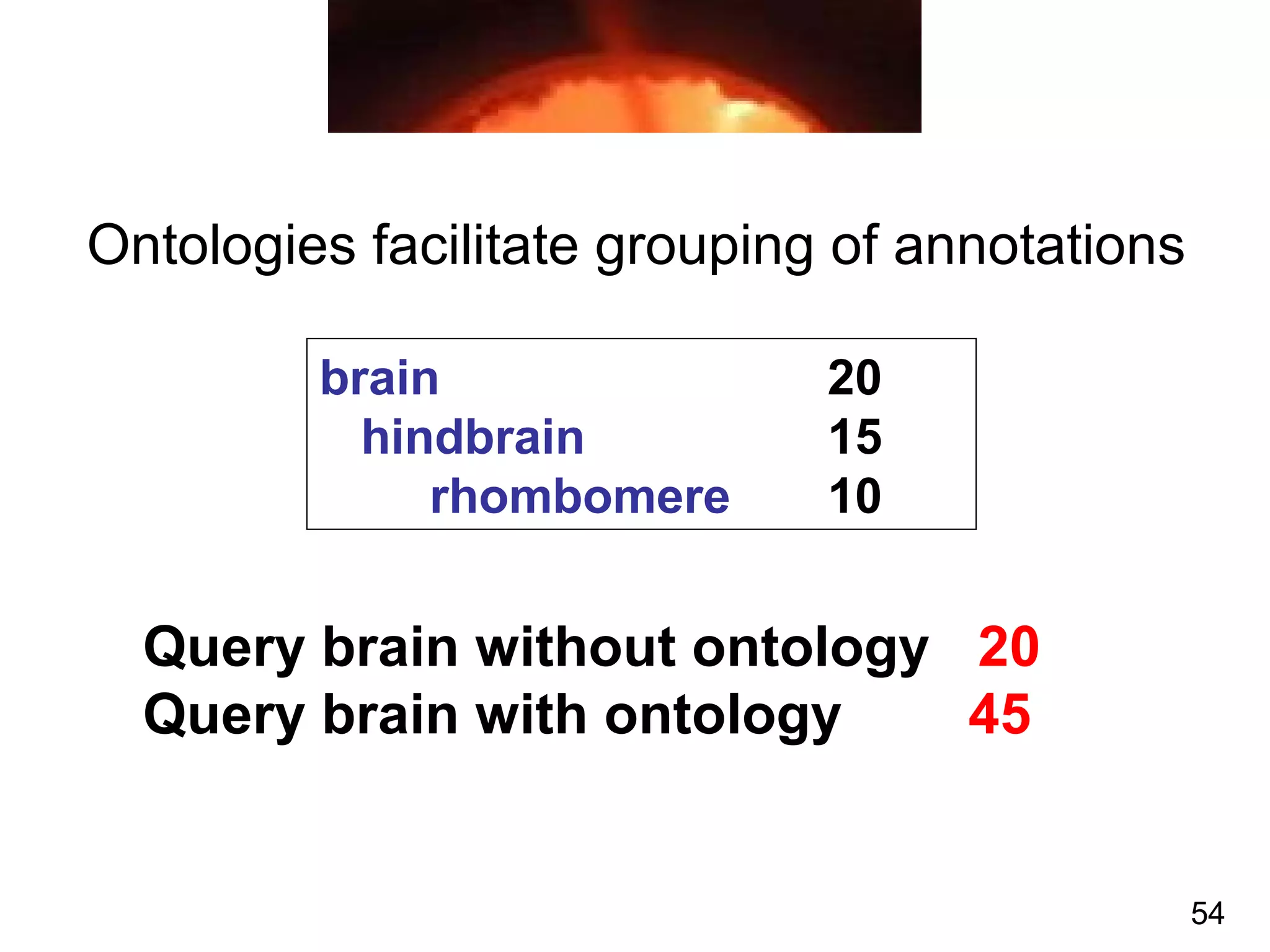
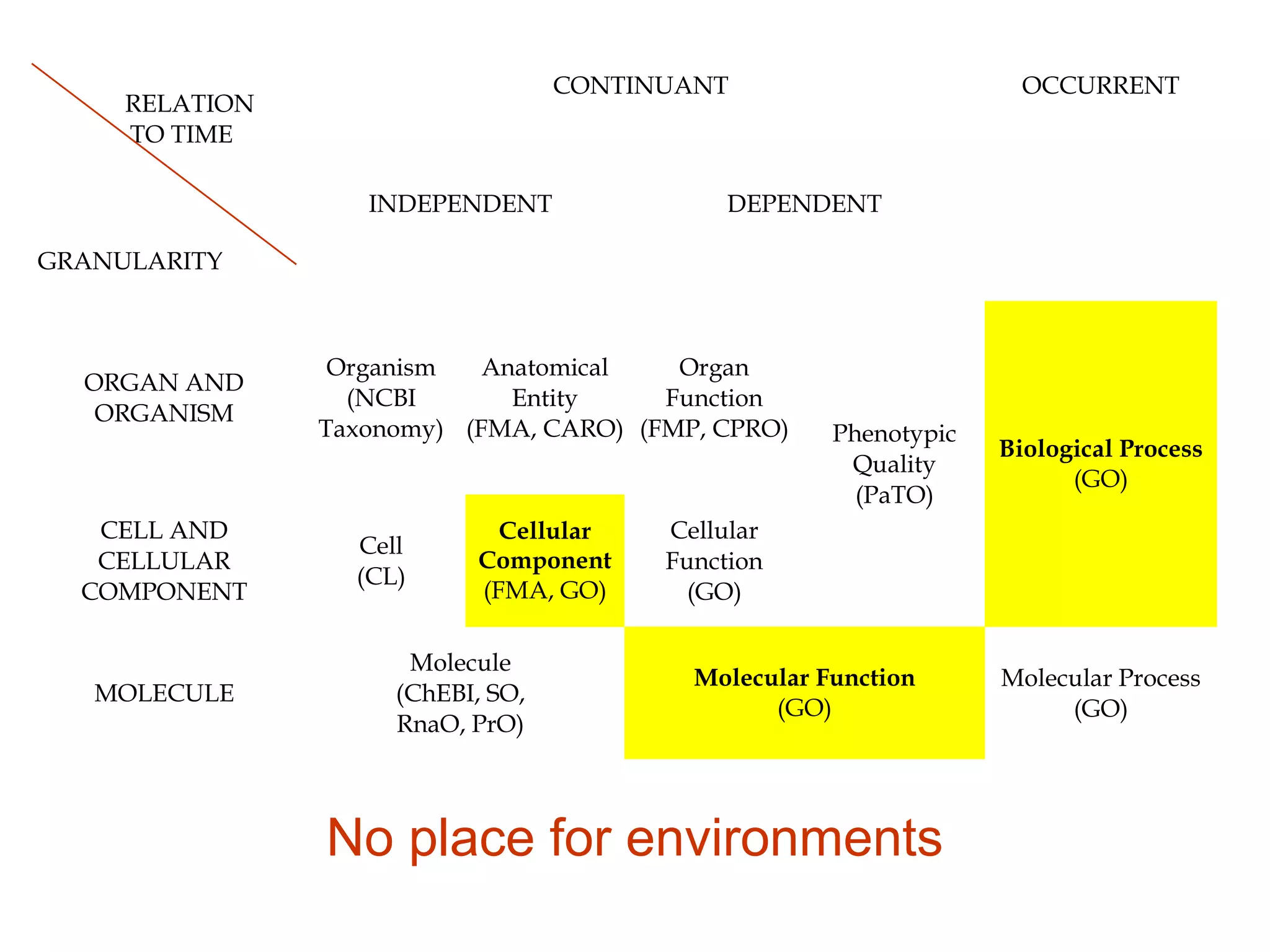
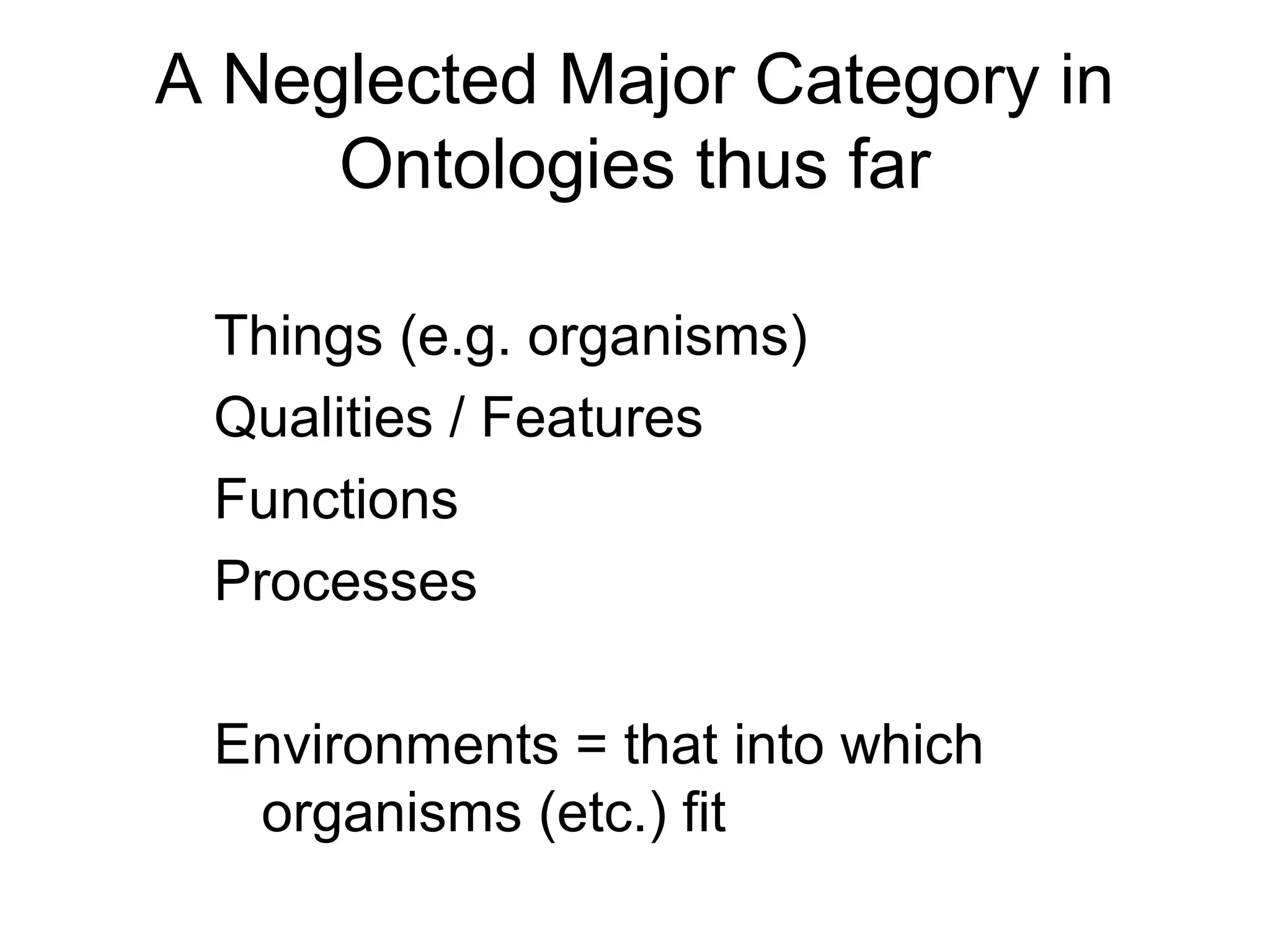
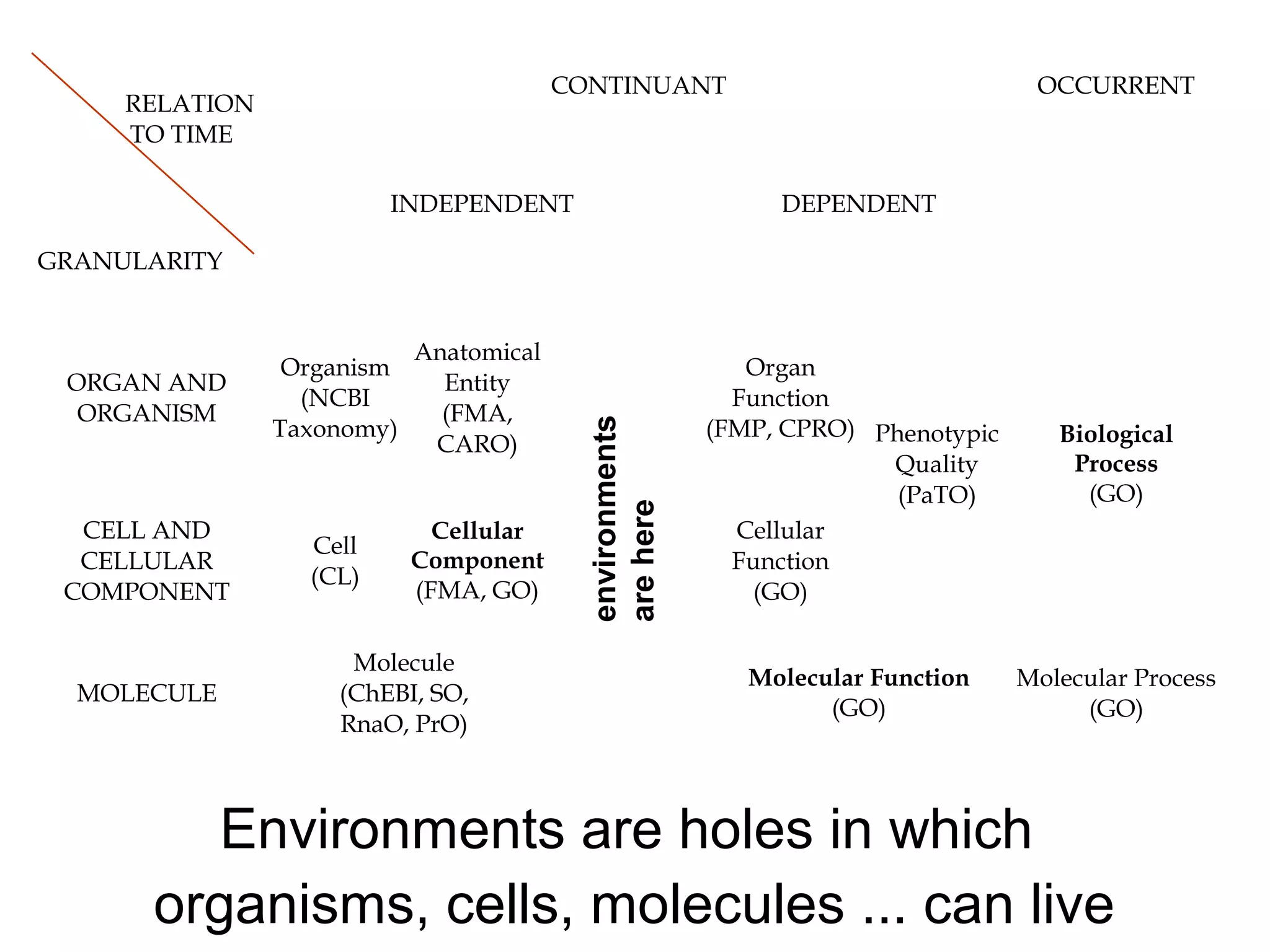
1. The document introduces ontologies for environmental biology and discusses several disciplines that could benefit from their use, including GIS, ecology, environmental biology, and various "-omics" fields.
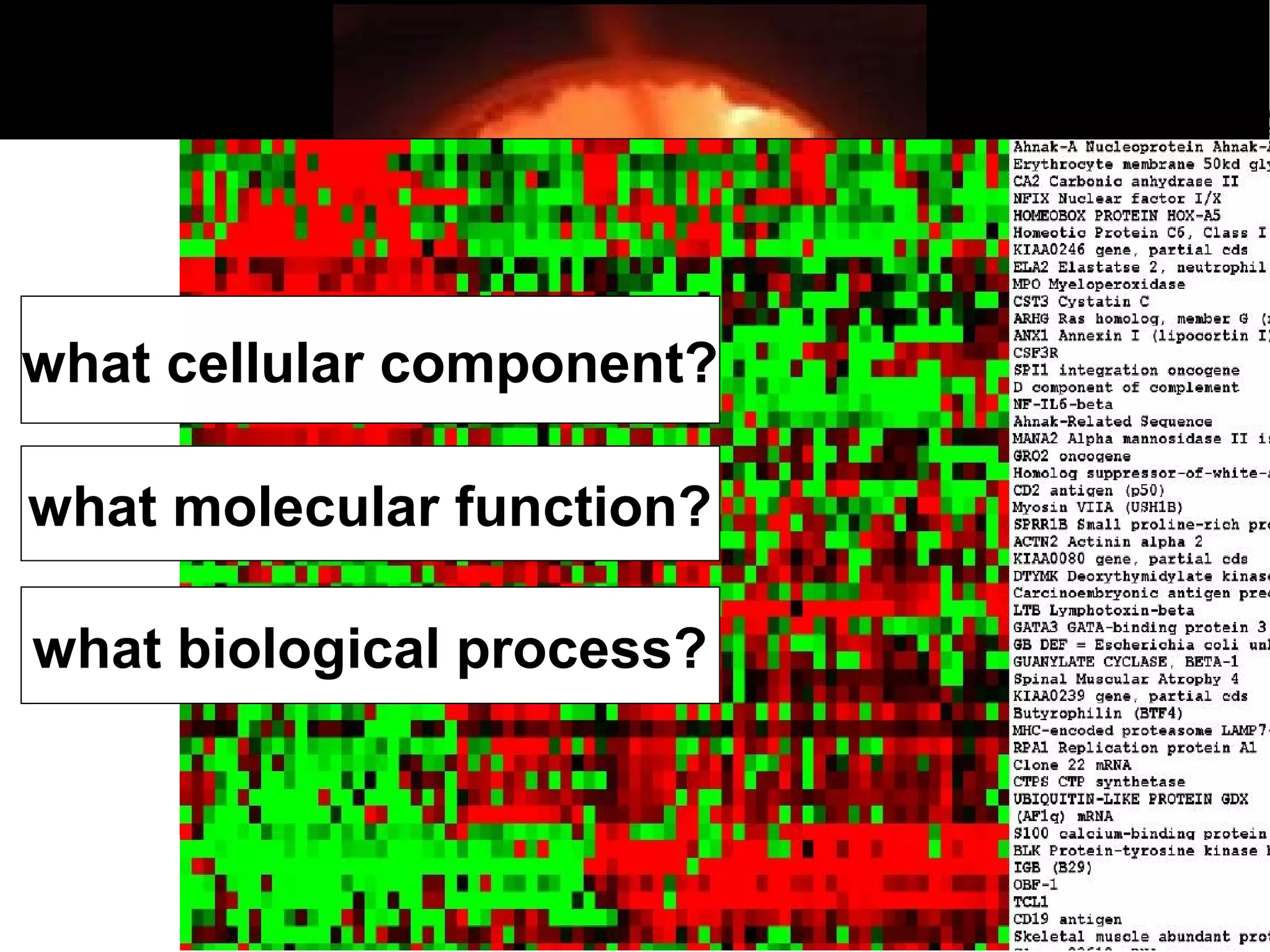
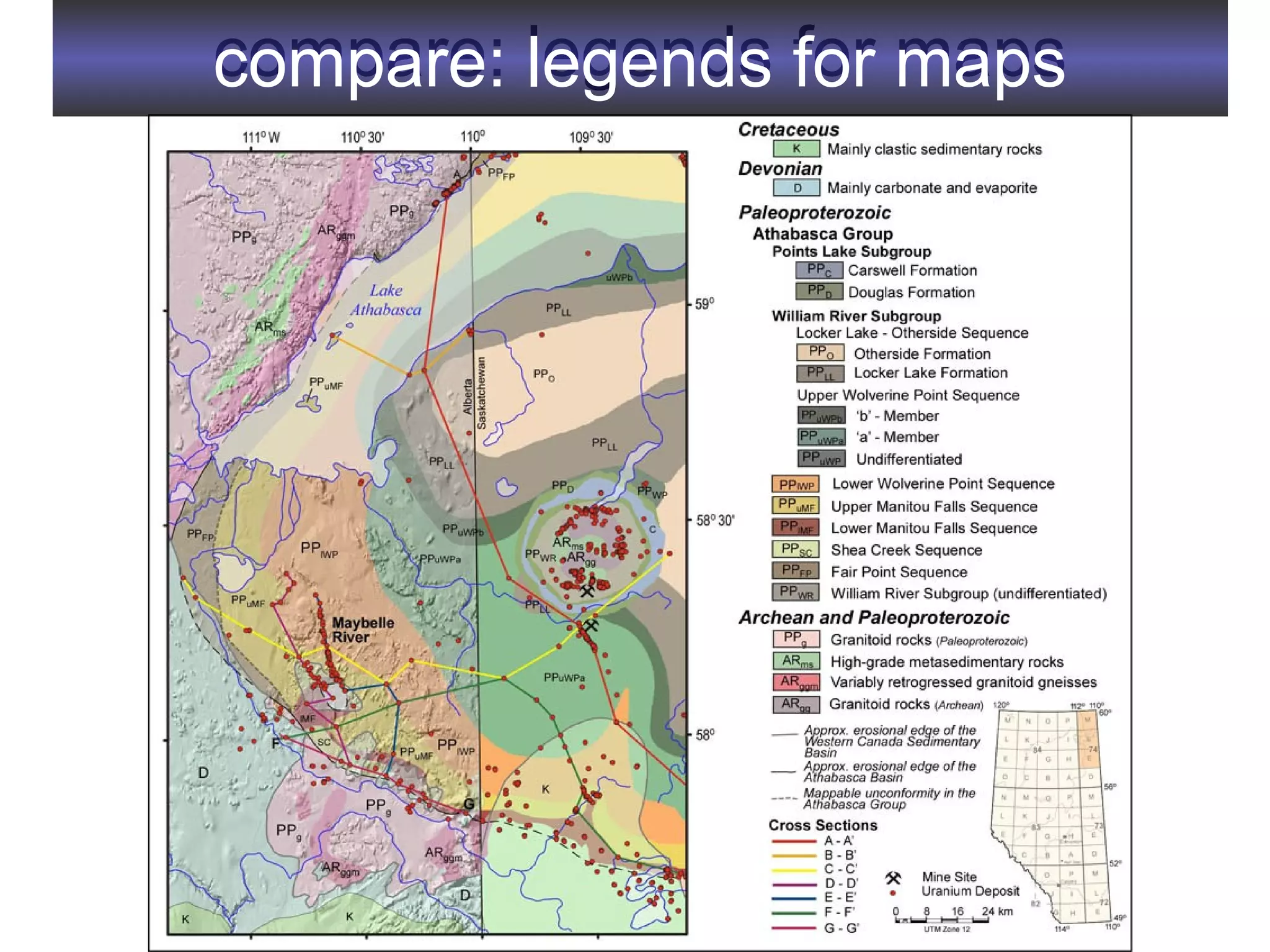
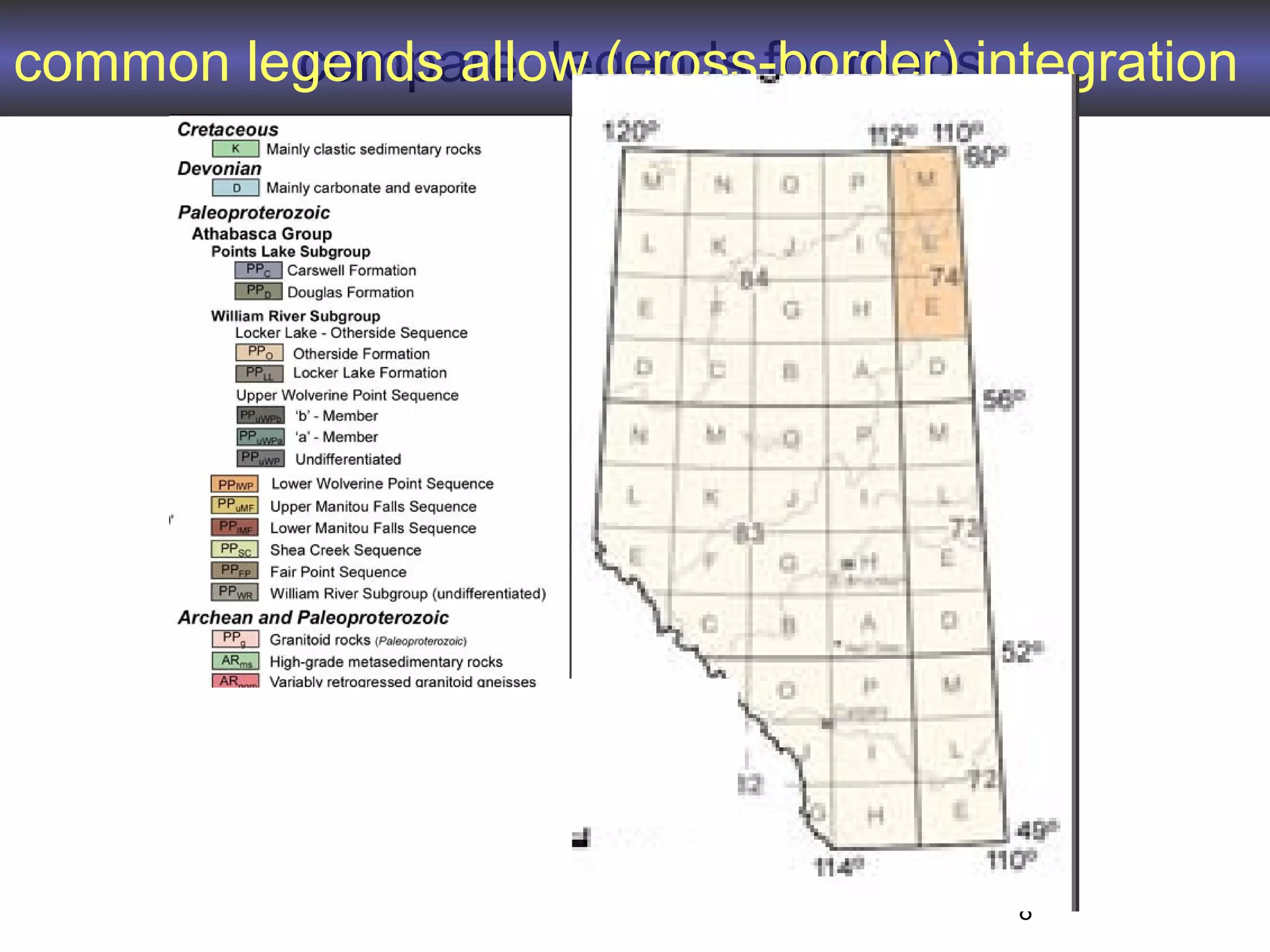
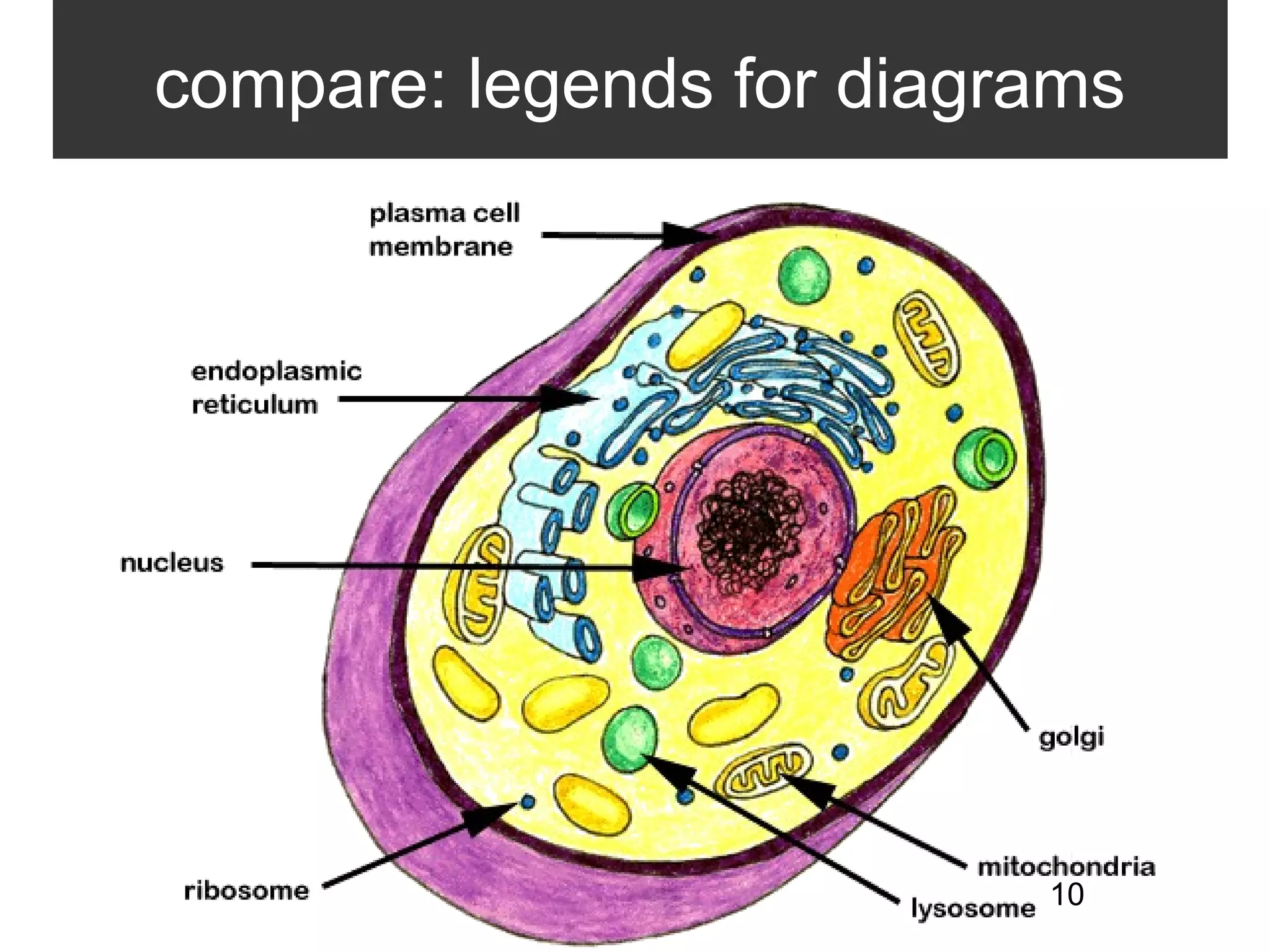
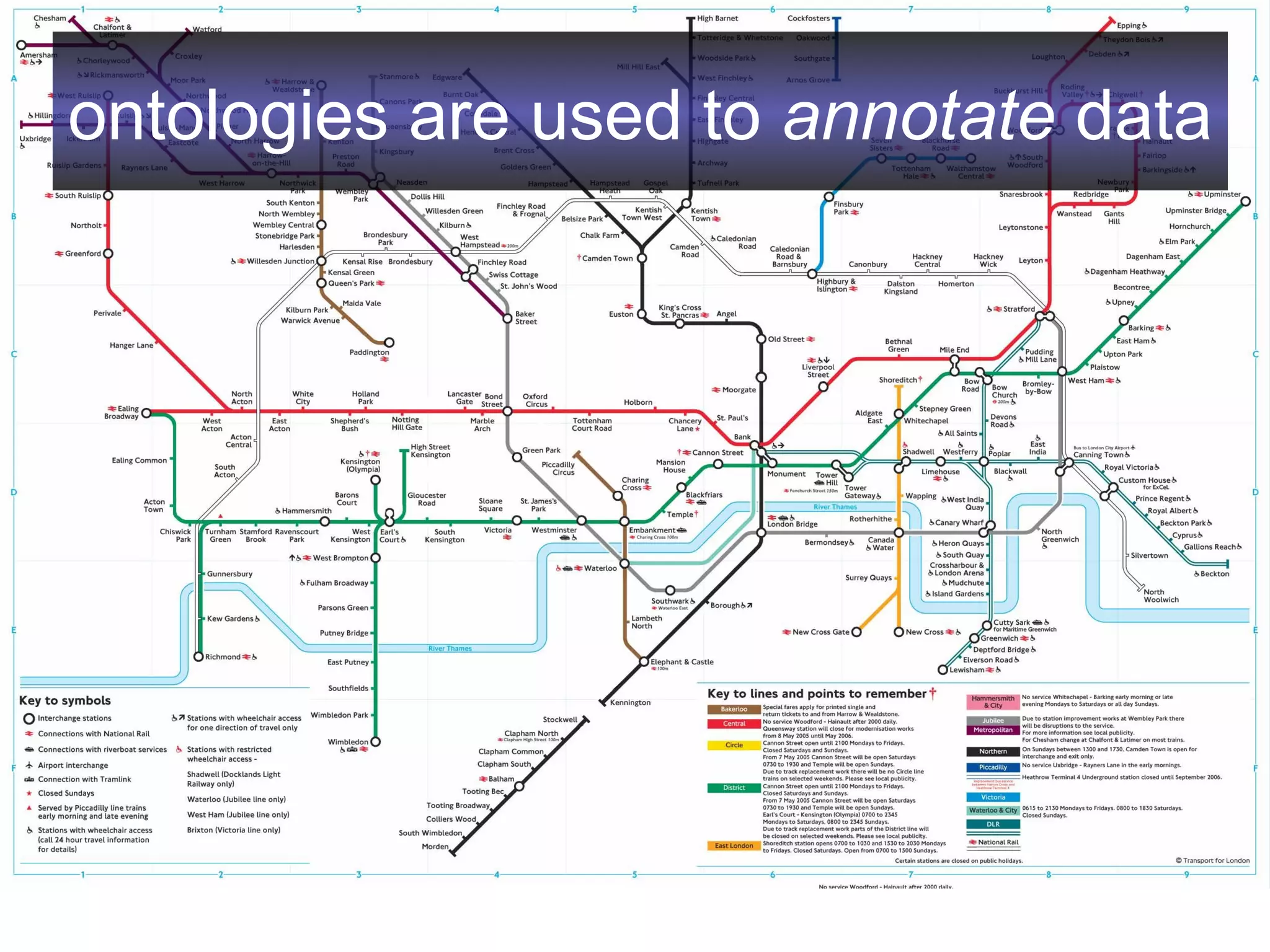
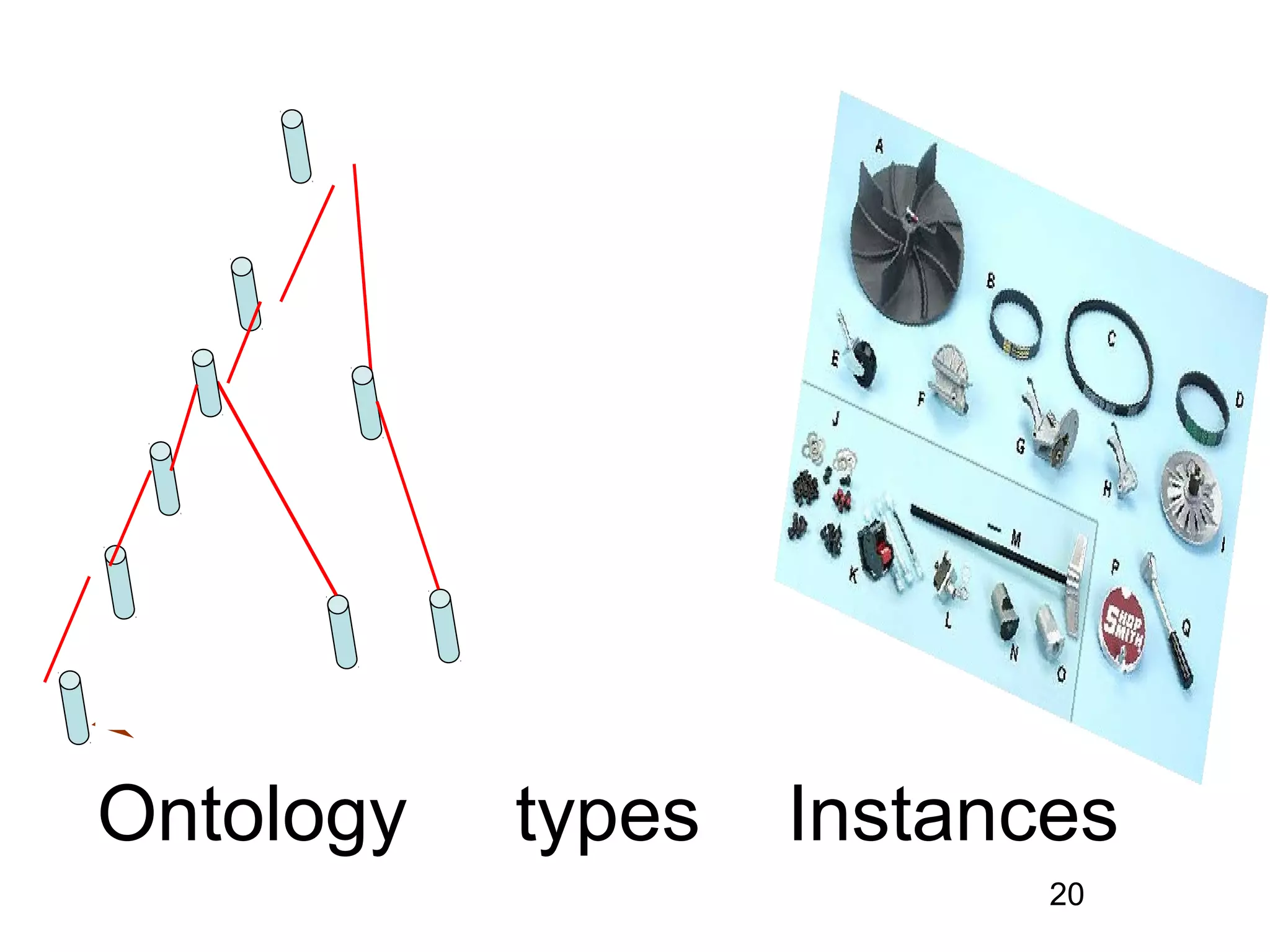
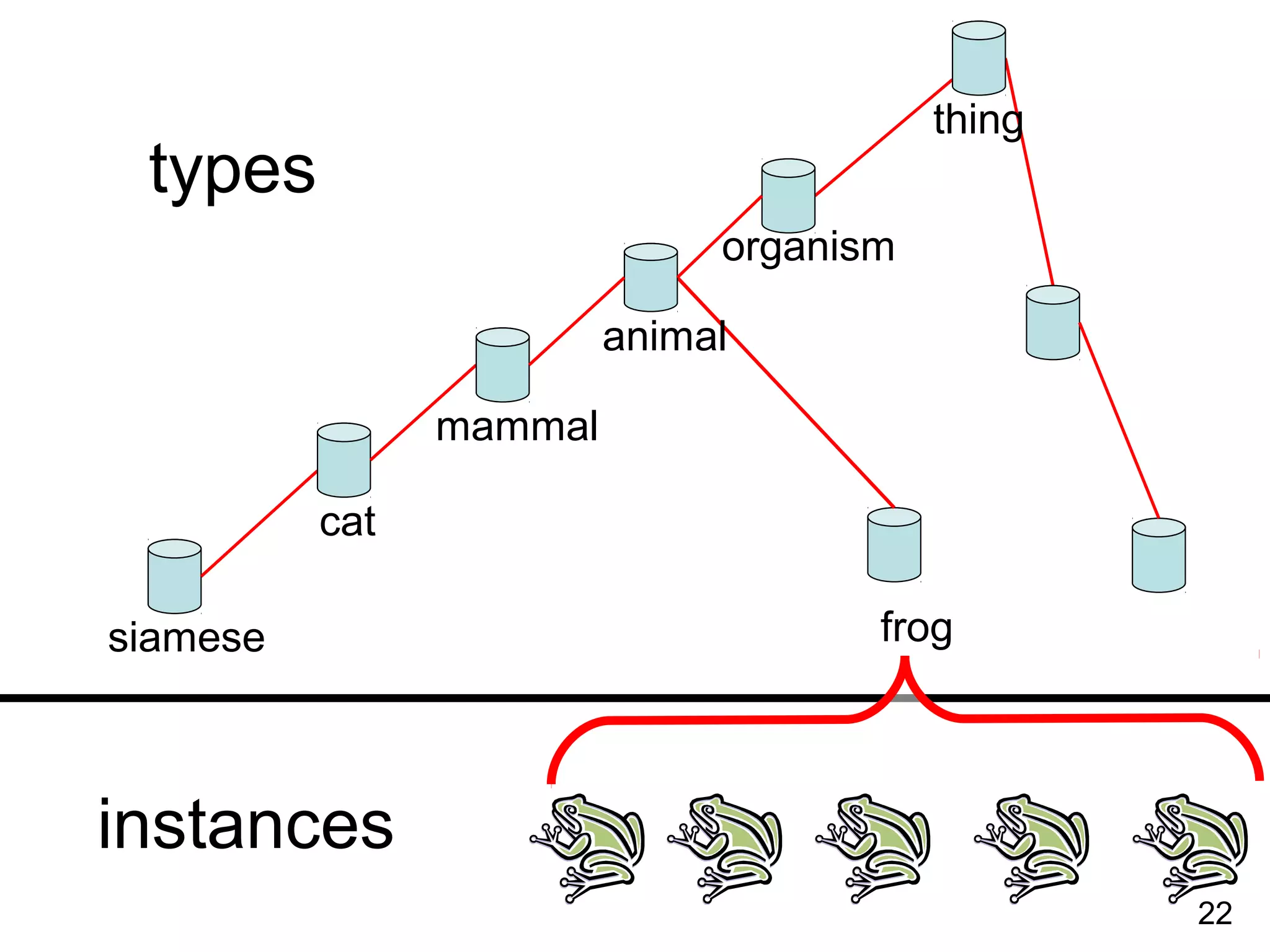
2. It describes what an ontology is and compares ontologies to legends for maps or diagrams, which allow integration and help humans and computers make sense of complex data. Ontologies provide standardized terminology and annotations.
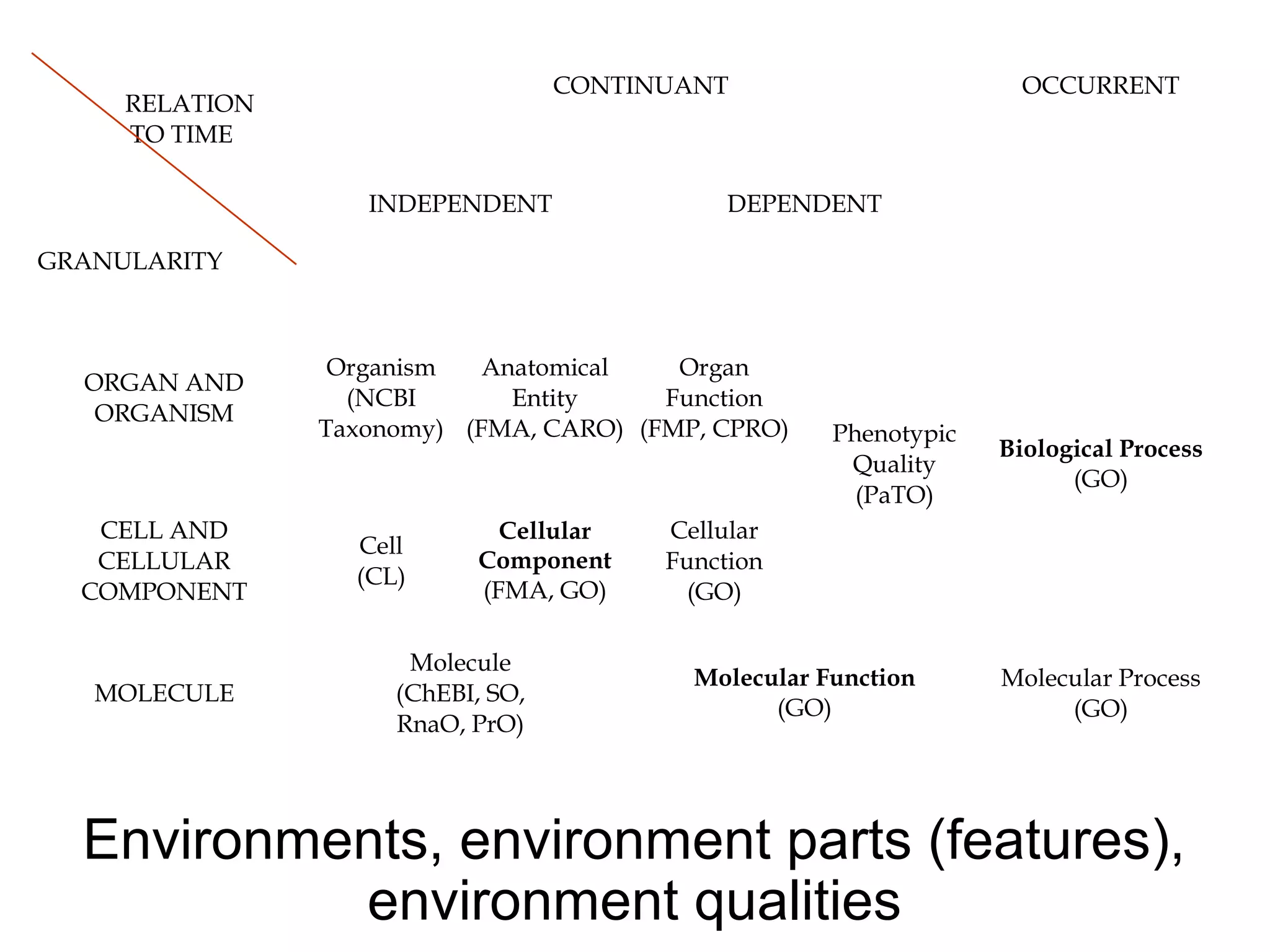
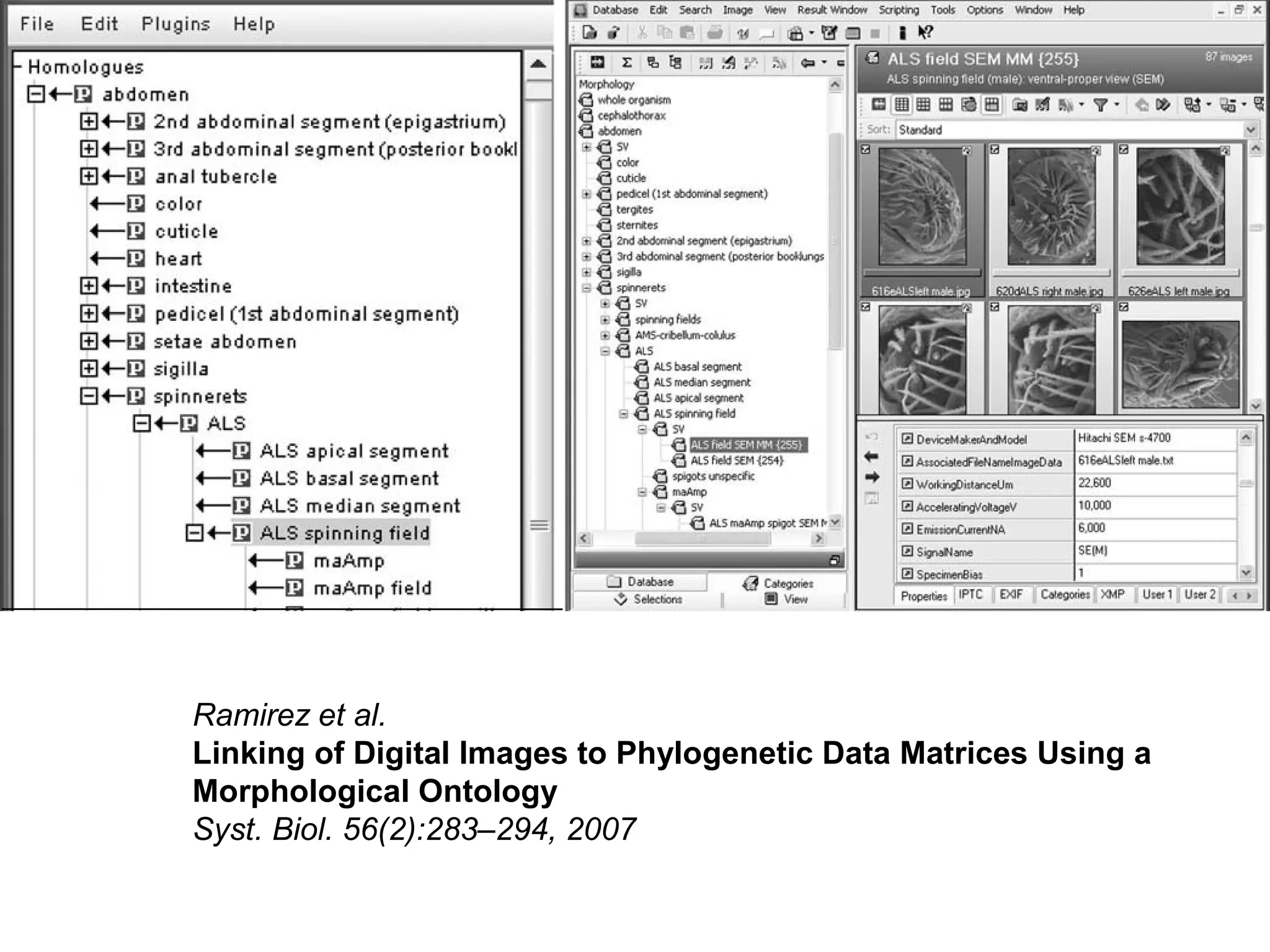
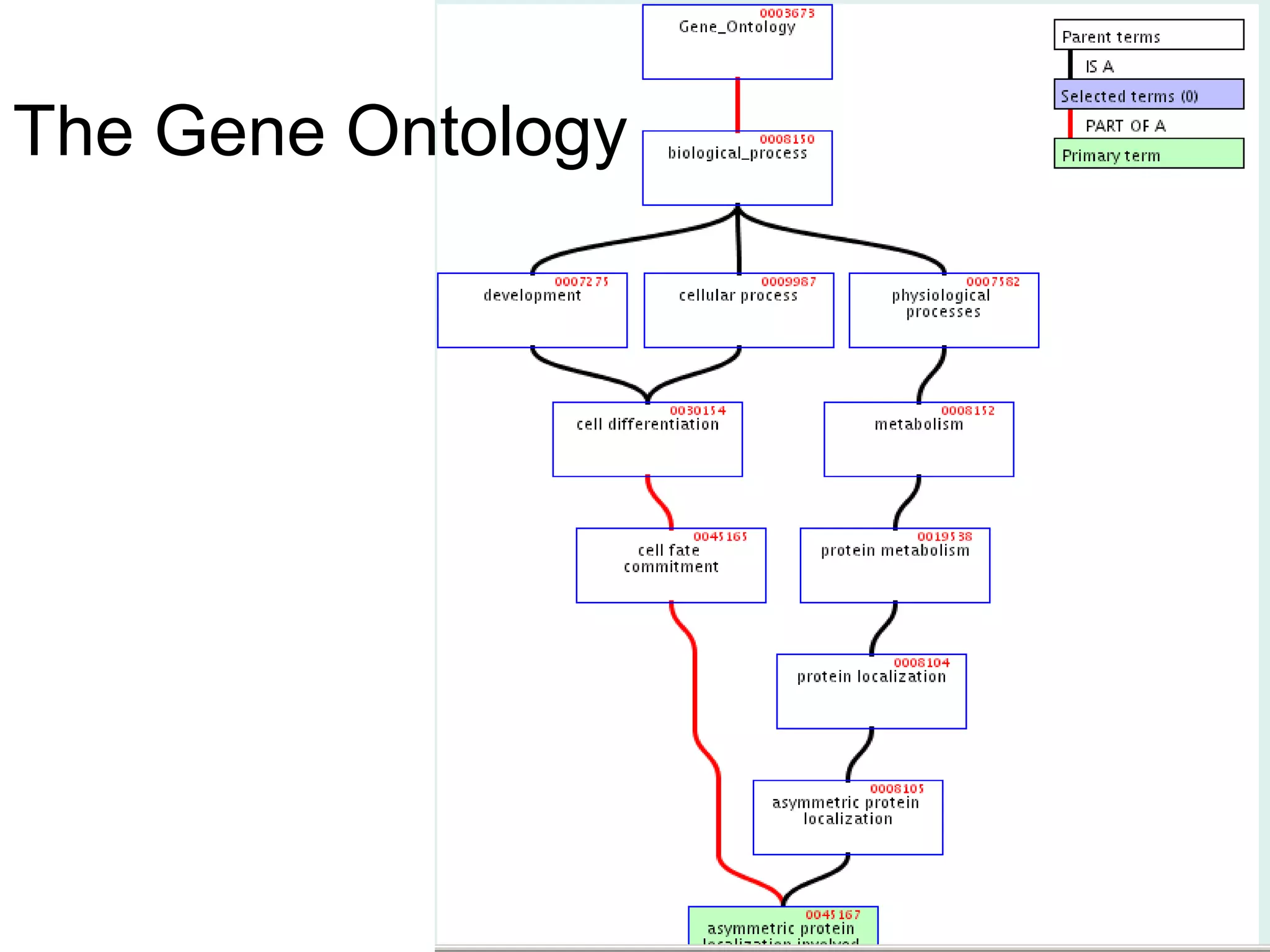
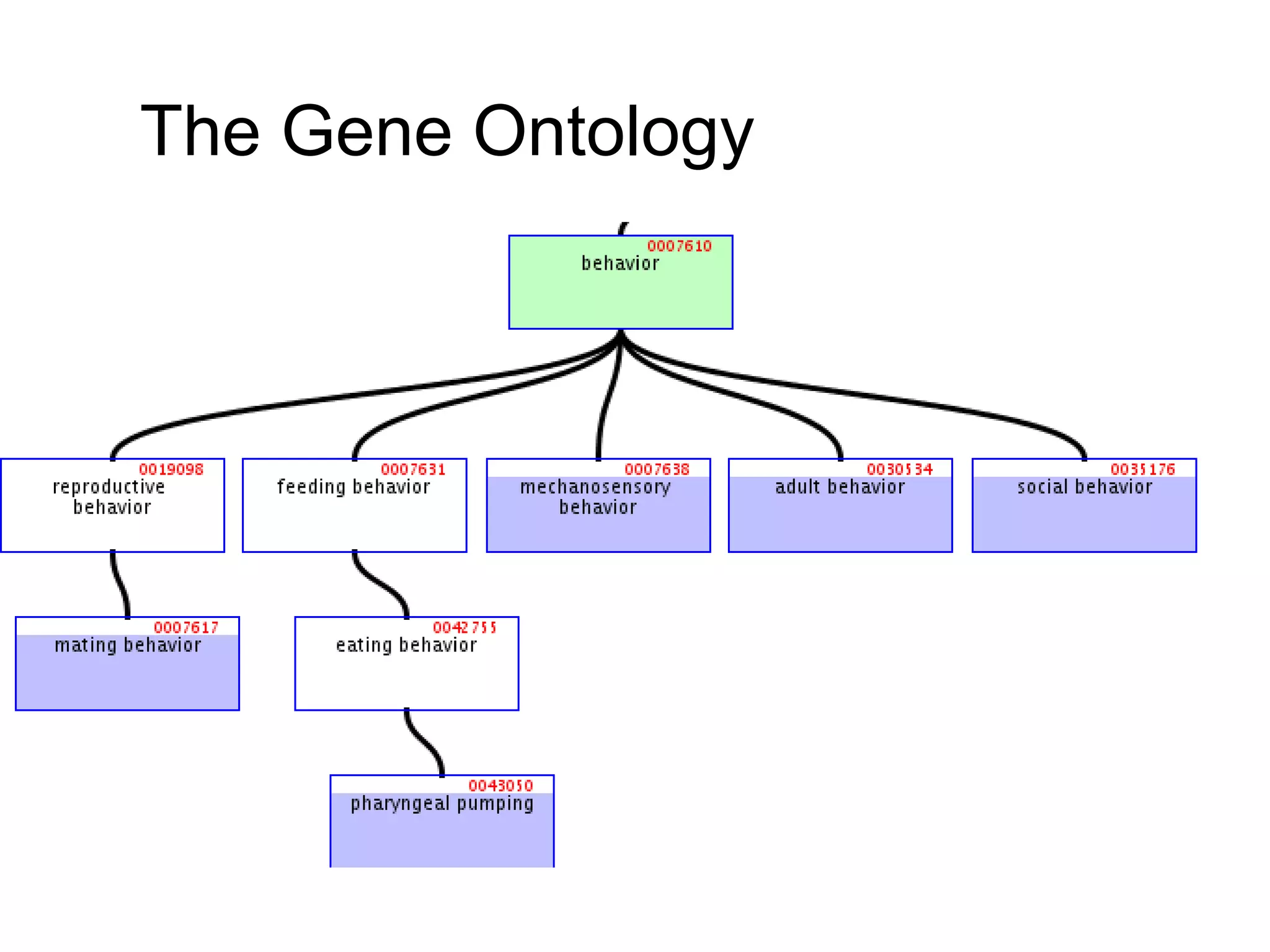
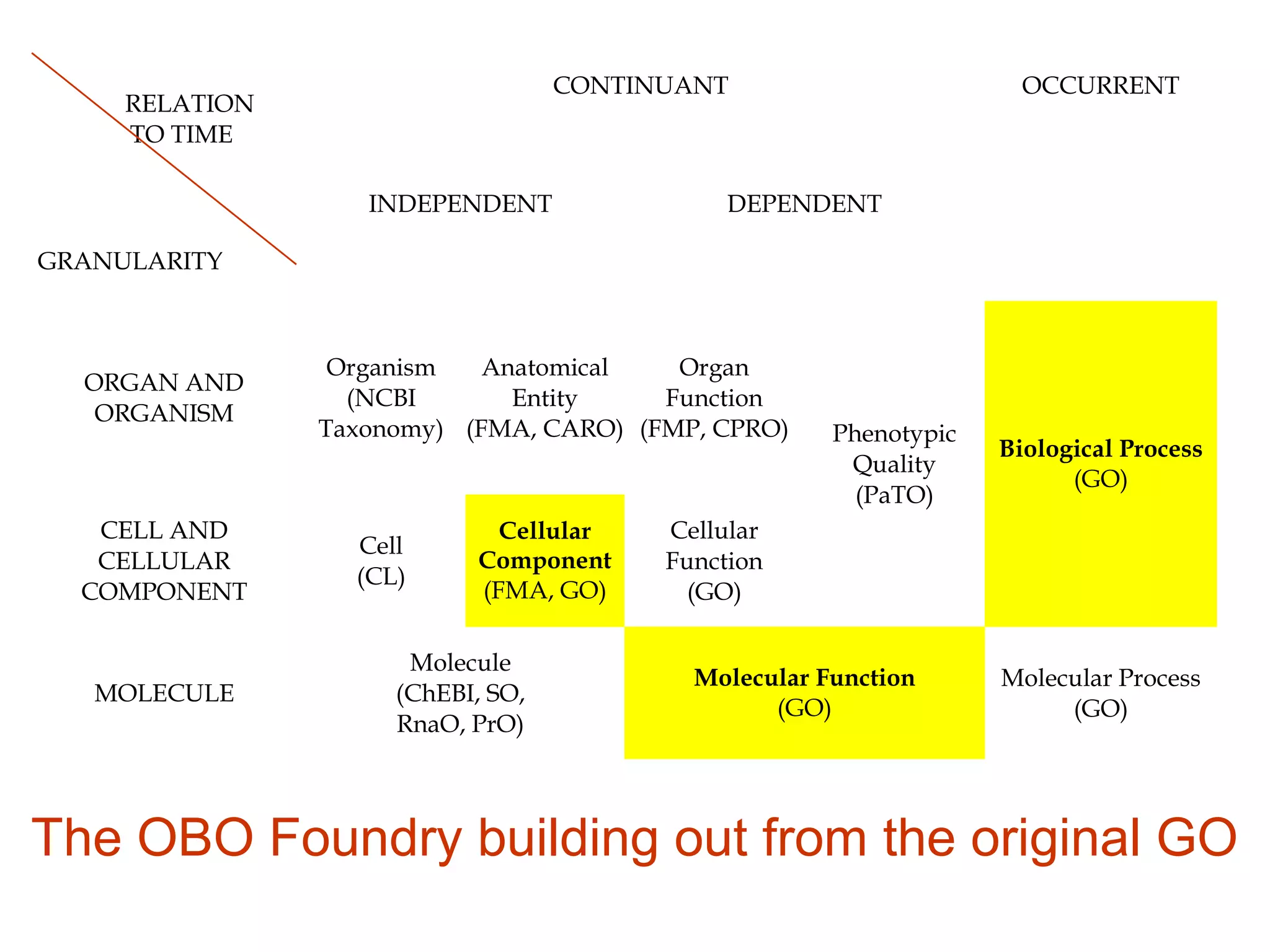
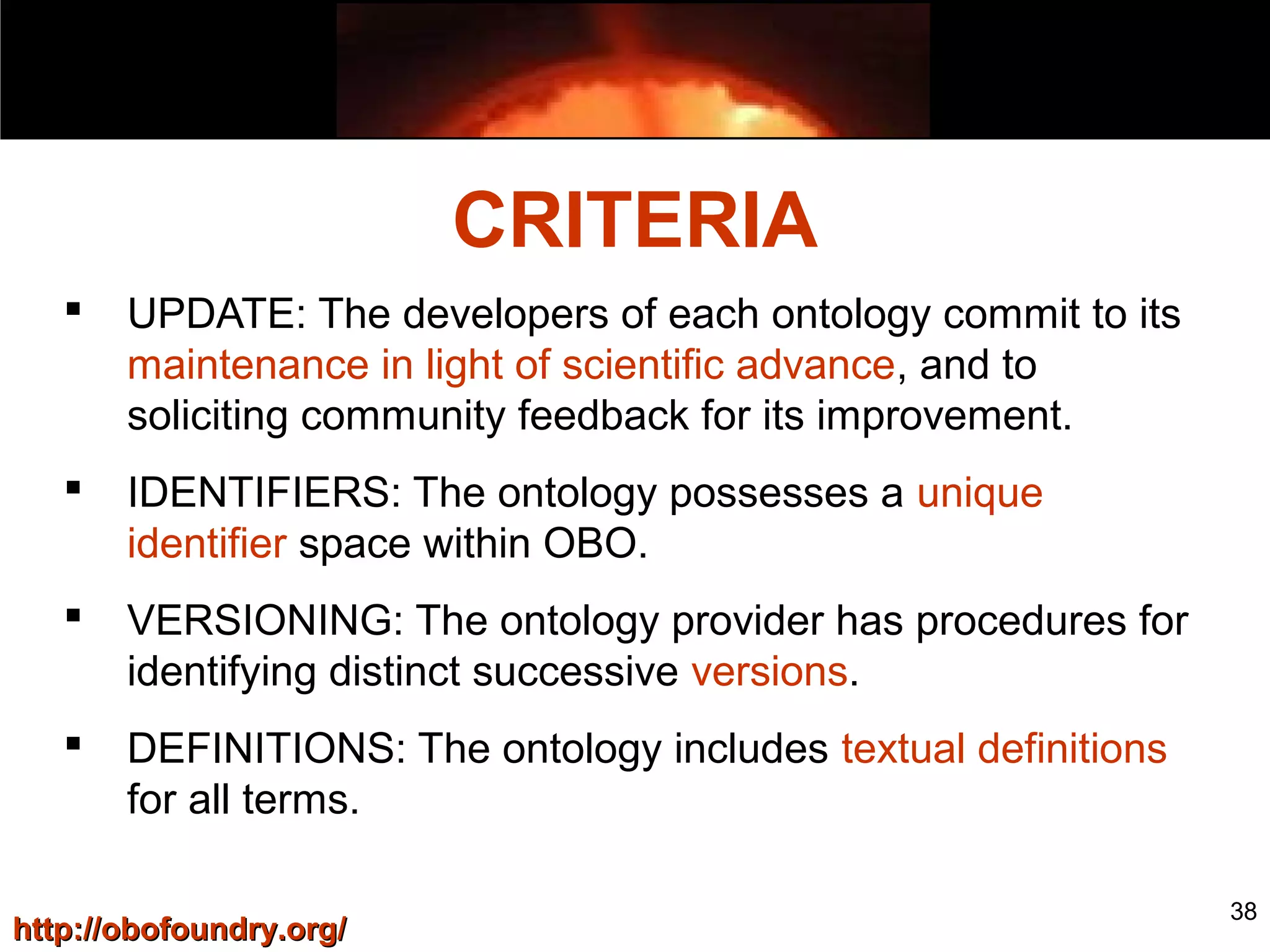
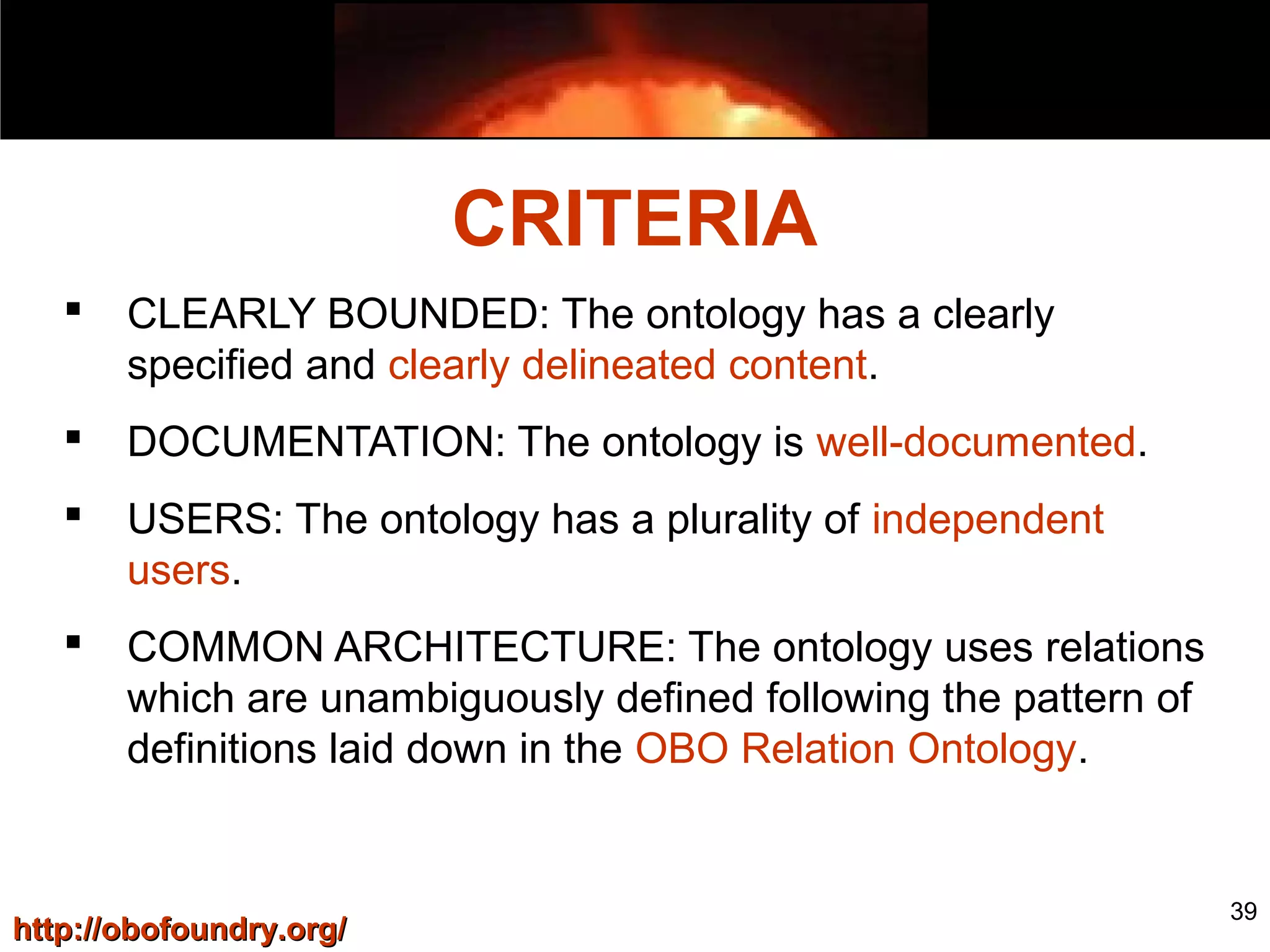
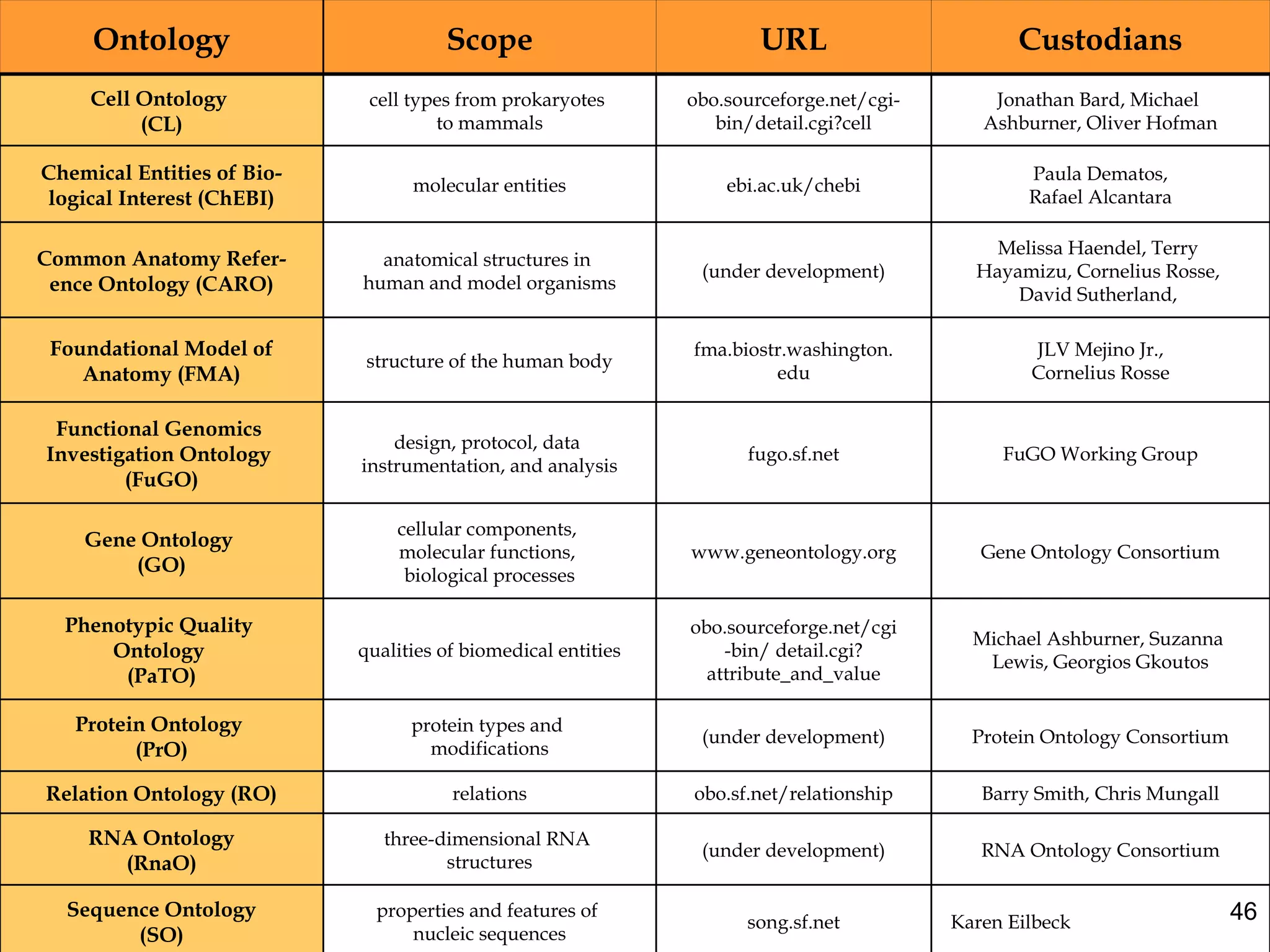
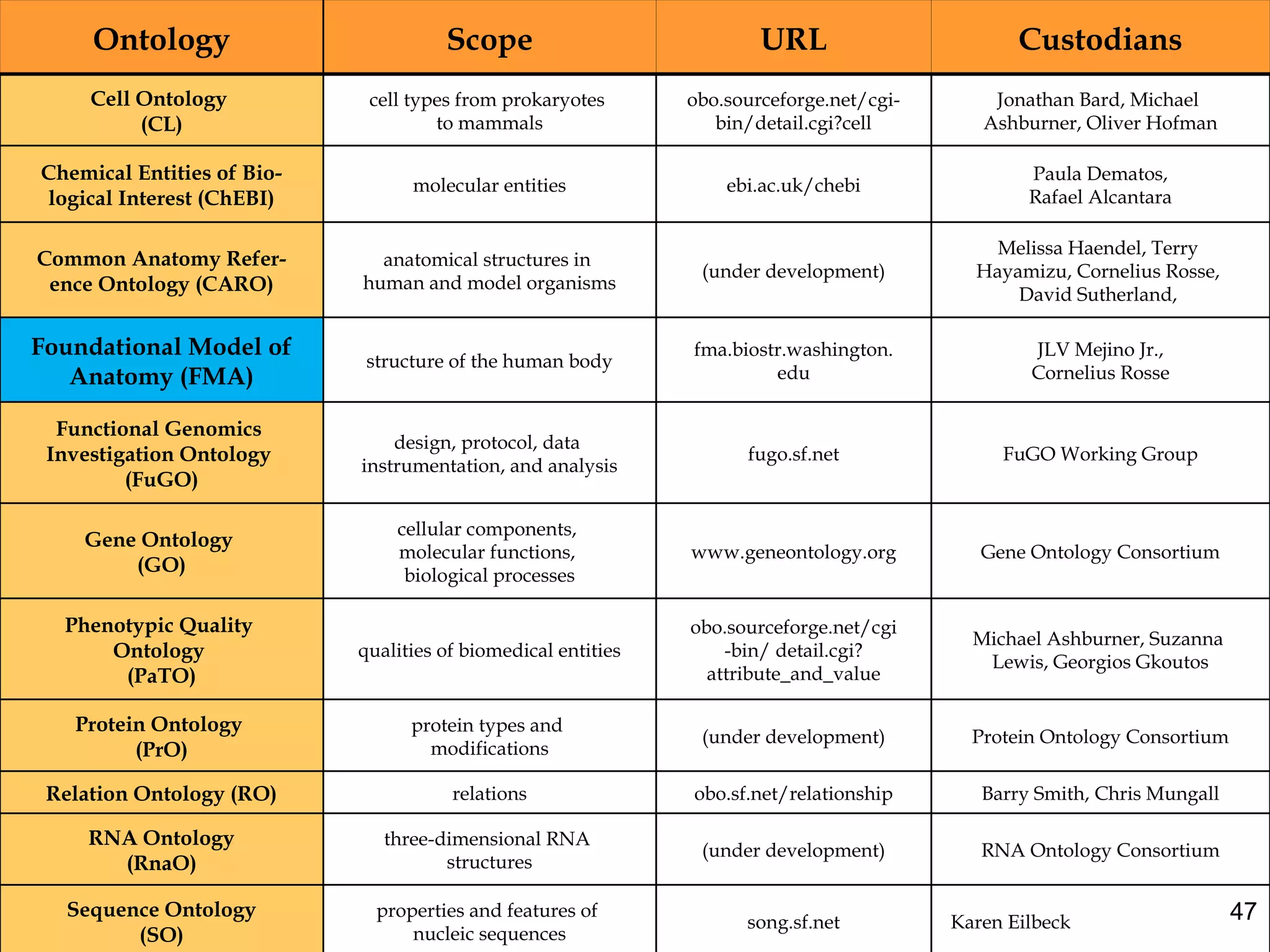
3. The document outlines the Open Biomedical Ontologies (OBO) Foundry, a collection of interoperable reference ontologies for annotating biomedical data. Foundry ontologies include the Gene Ontology and other ontologies for molecules, cells, anatomical structures, and more. They are developed through consensus and share





































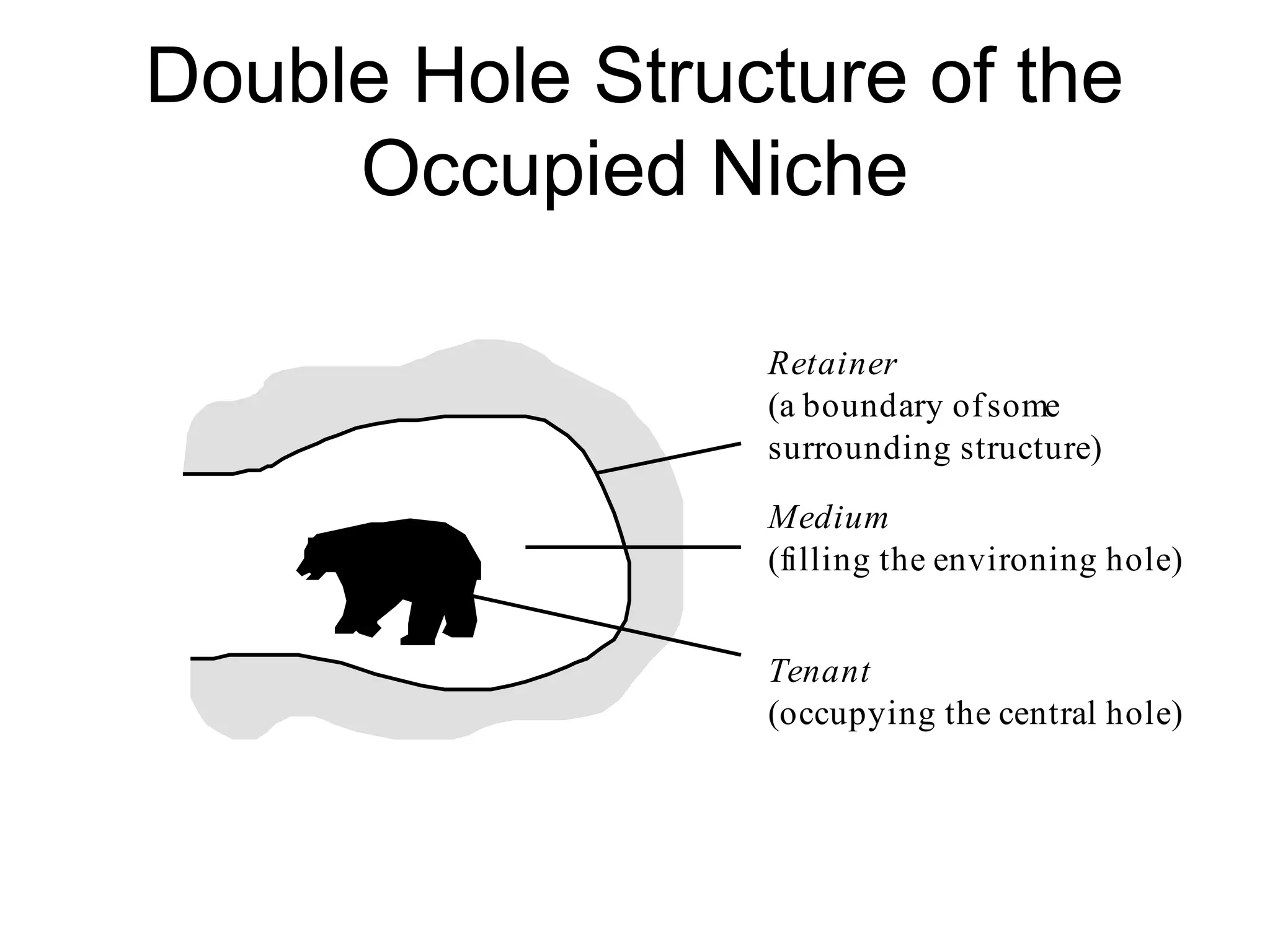






![Elton – niche as role
the ‘niche’ of an animal means
its place in the biotic environment, its
relations to food and enemies. [...]
When an ecologist says ‘there goes a
badger’ he should include in his thoughts
some definite idea of the animal’s place in
the community to which it belongs,
just as if he had said ‘there goes the vicar’
(Elton 1927, pp. 63f.)](https://image.slidesharecdn.com/nichesoxfordaug07-130524145720-phpapp01/75/Introduction-to-Ontologies-for-Environmental-Biology-84-2048.jpg)





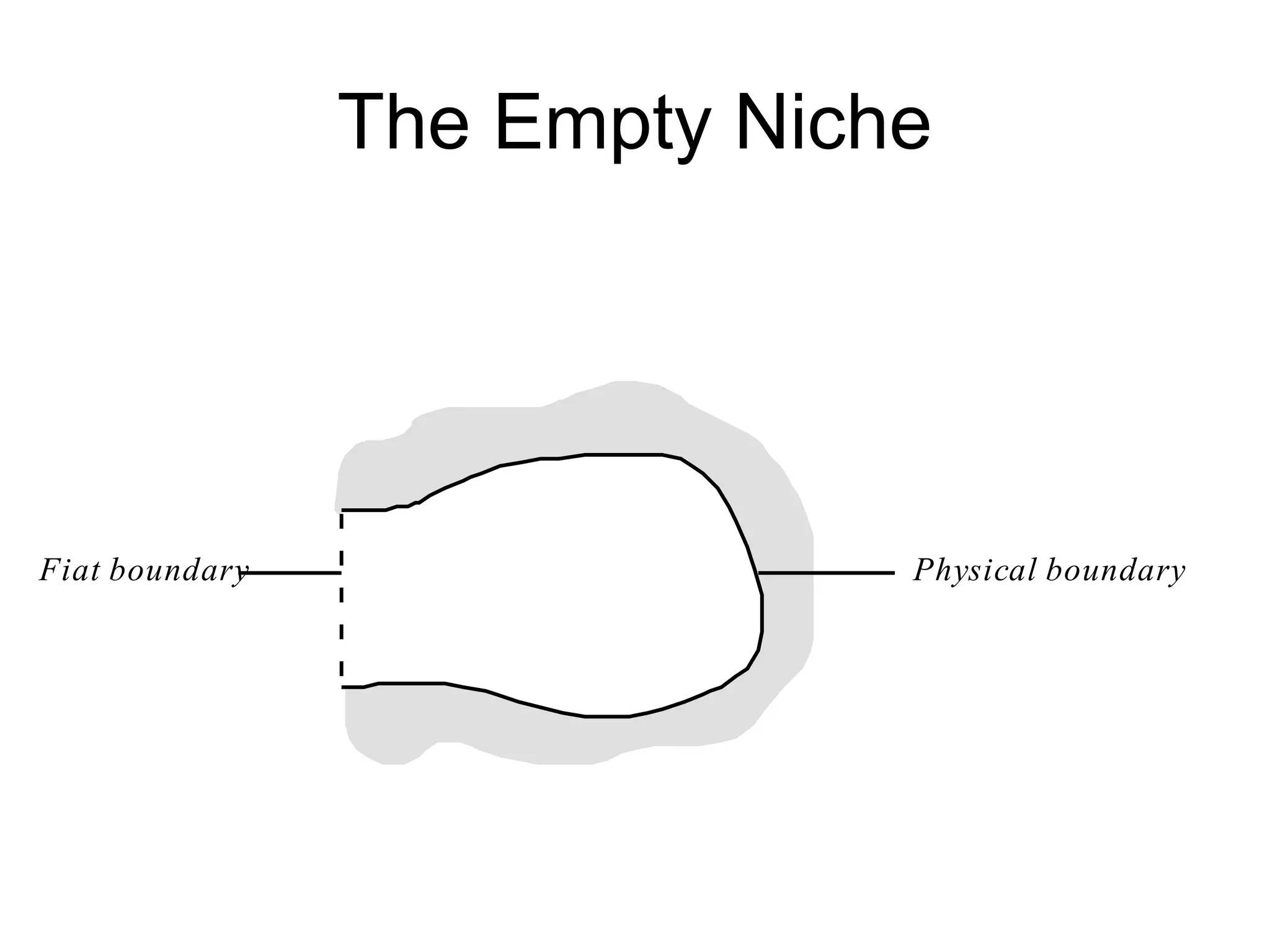
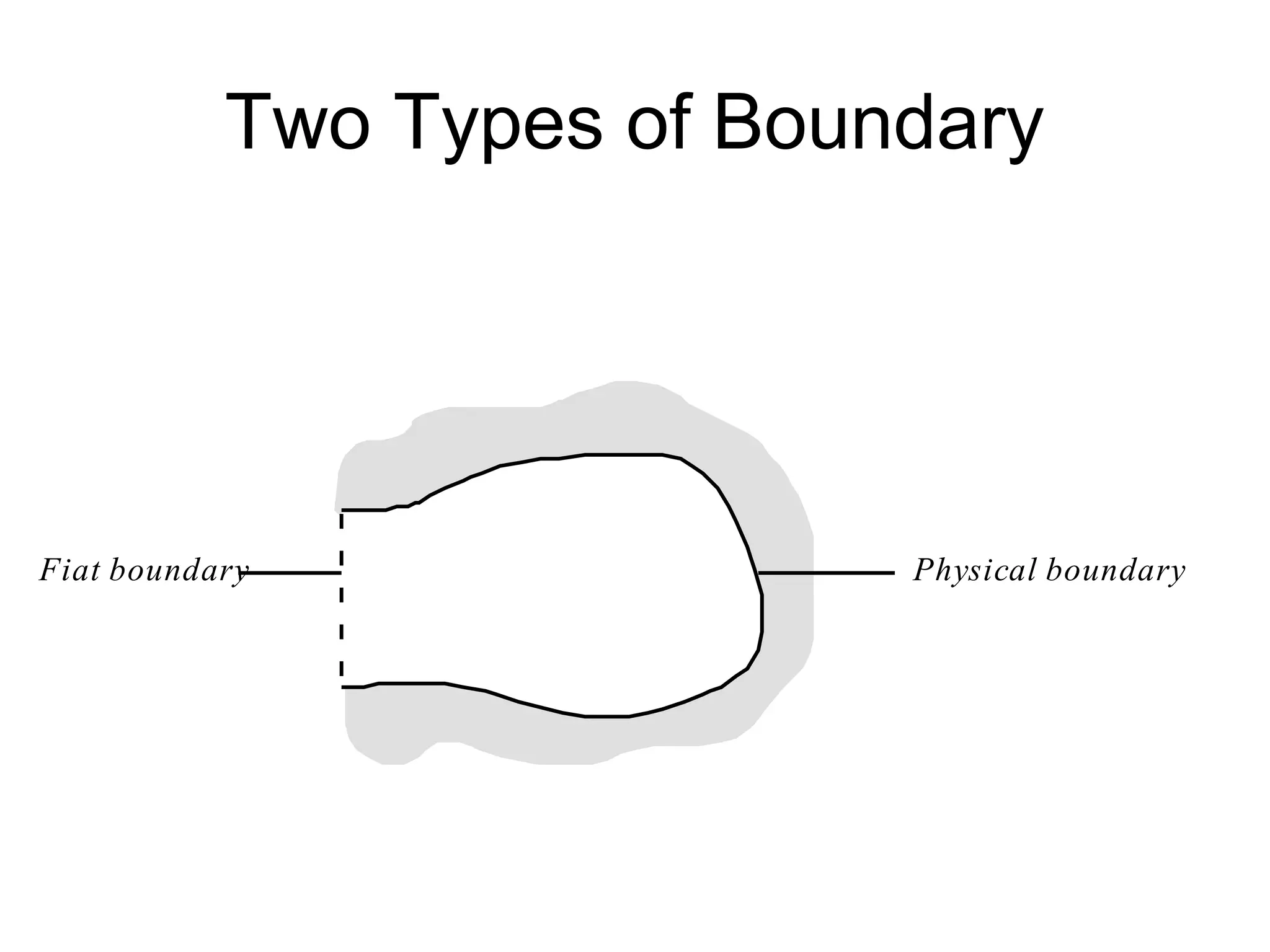
![J. J. Gibson’s Ecological
Psychology
The terrestrial environment is [best]
described in terms of a medium,
substances, and the surfaces that
separate them. (Gibson 1979, p. 16)](https://image.slidesharecdn.com/nichesoxfordaug07-130524145720-phpapp01/75/Introduction-to-Ontologies-for-Environmental-Biology-92-2048.jpg)