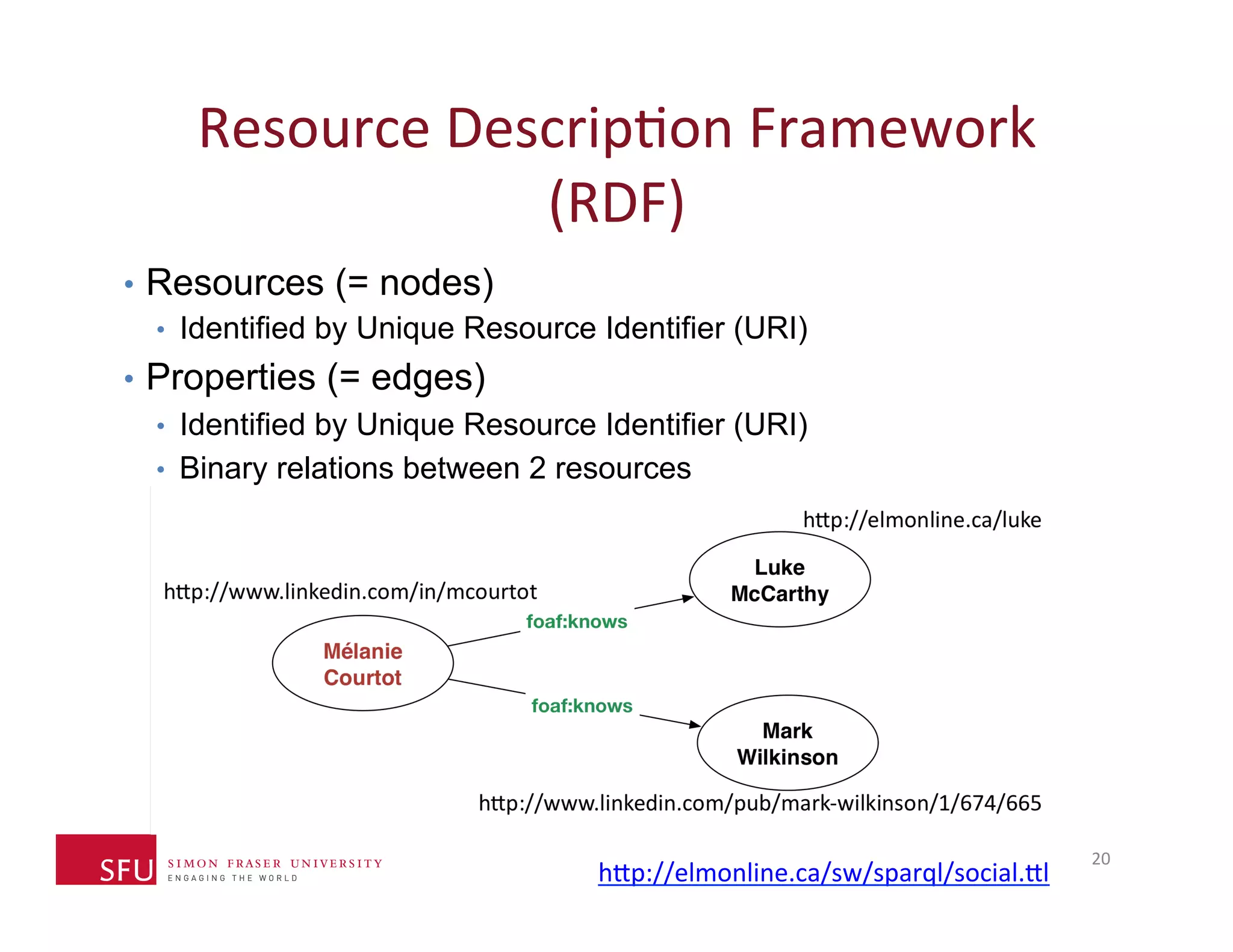
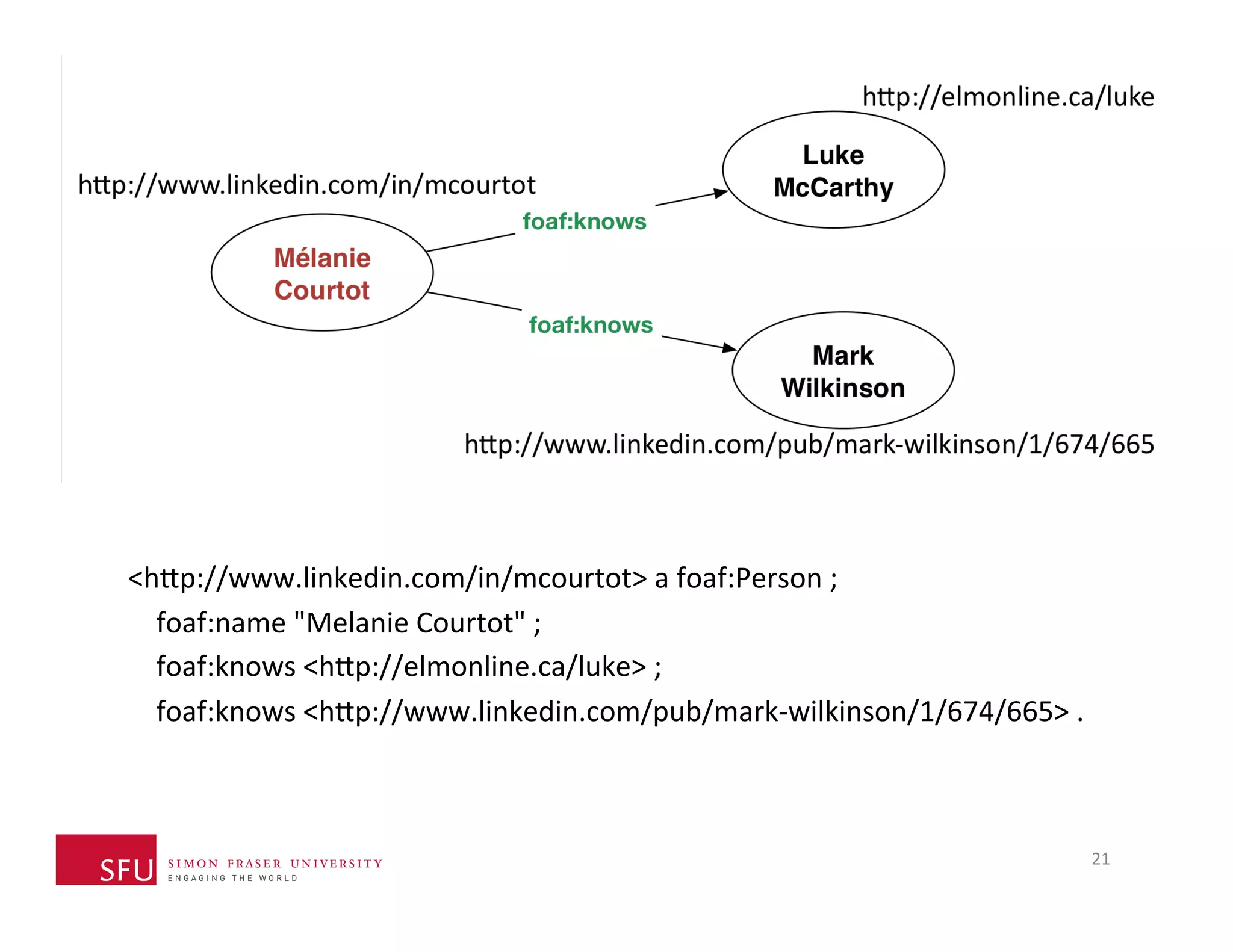
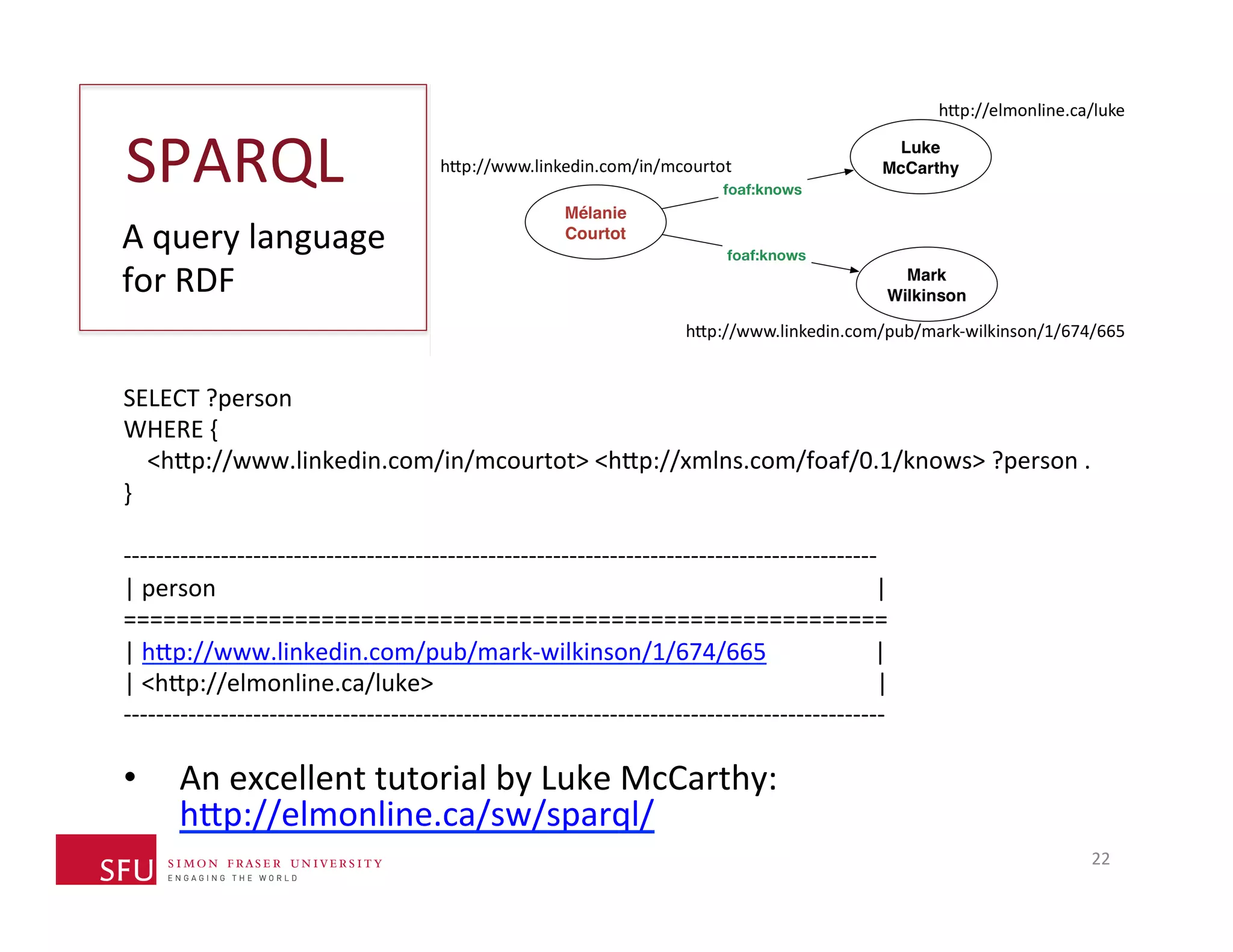
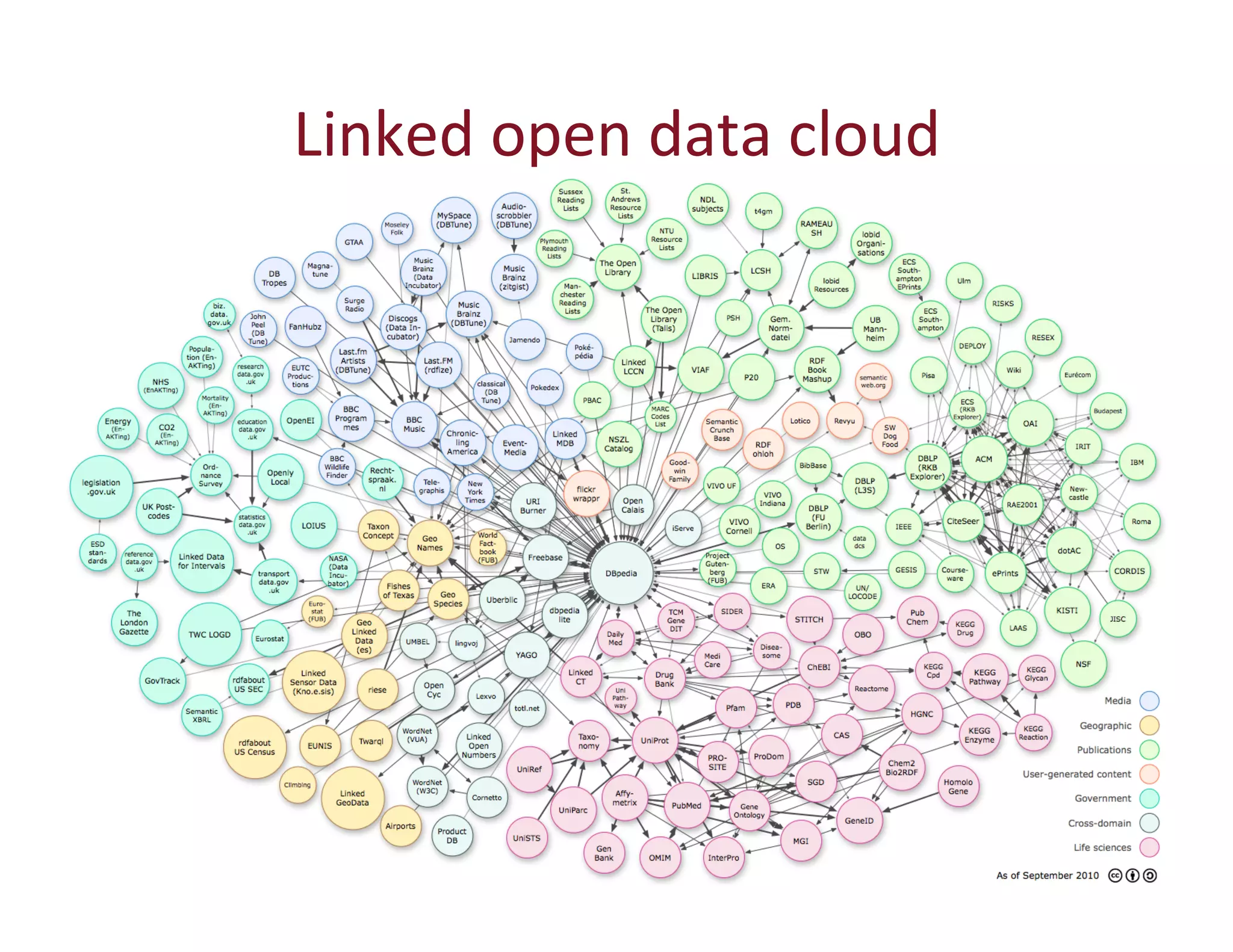
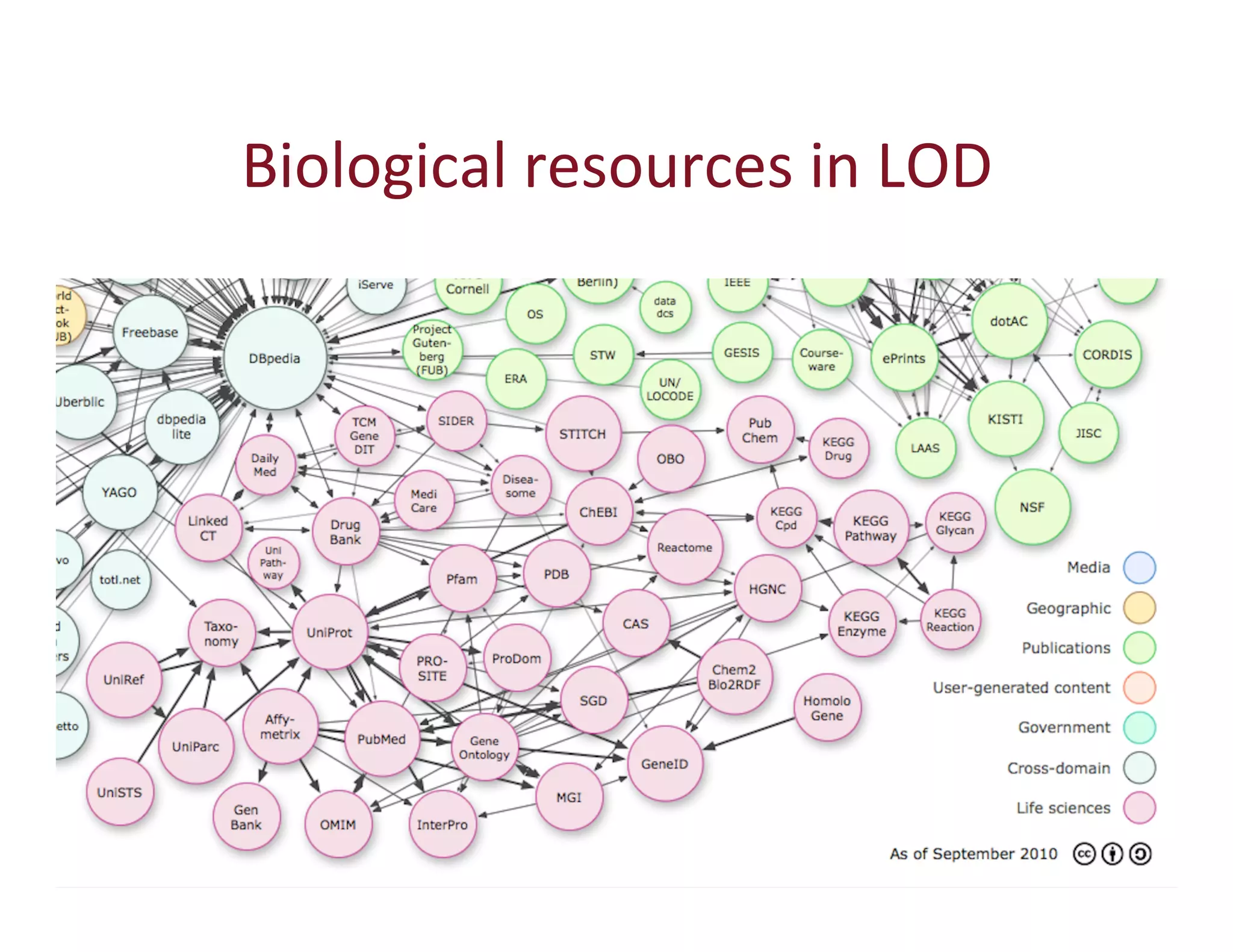
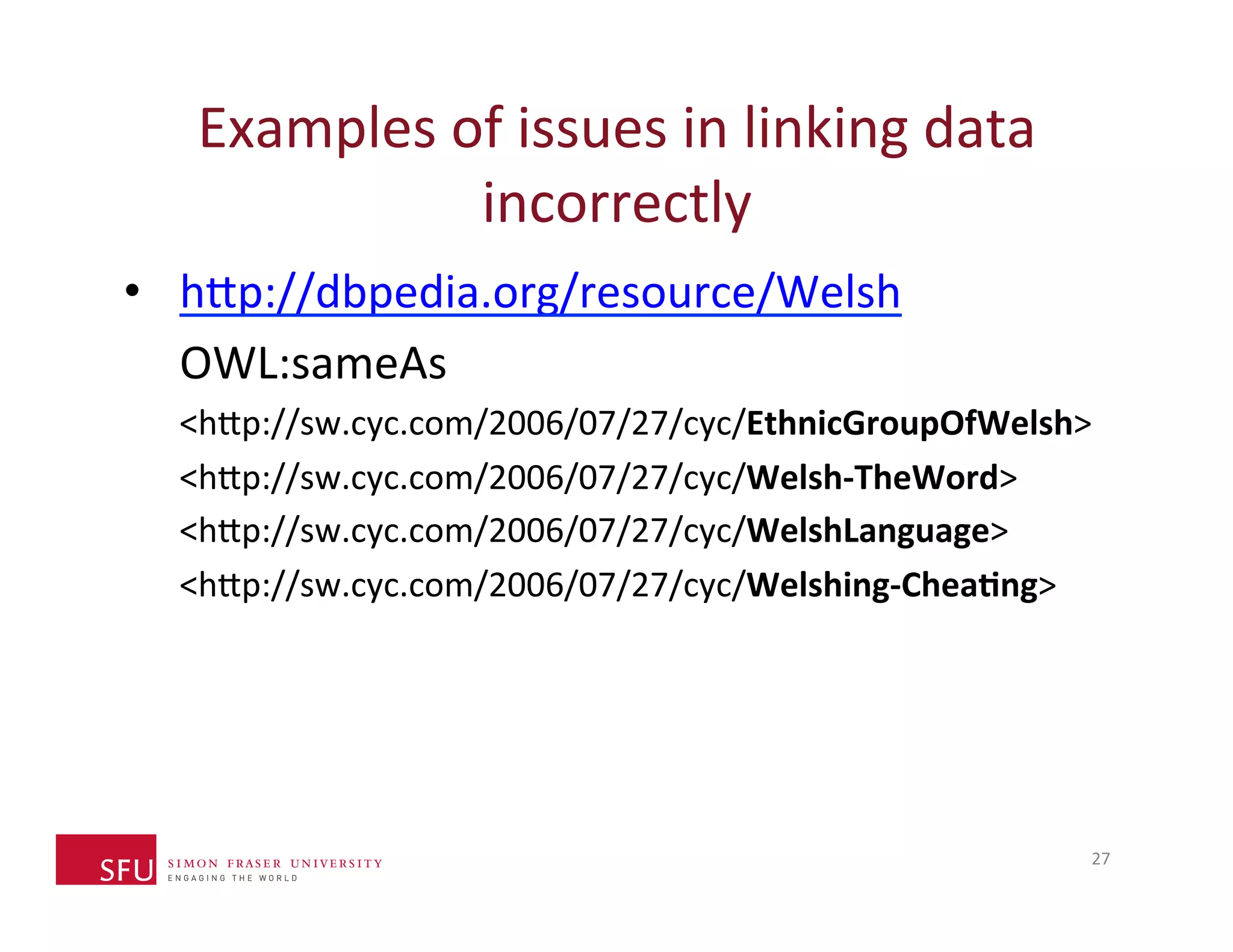
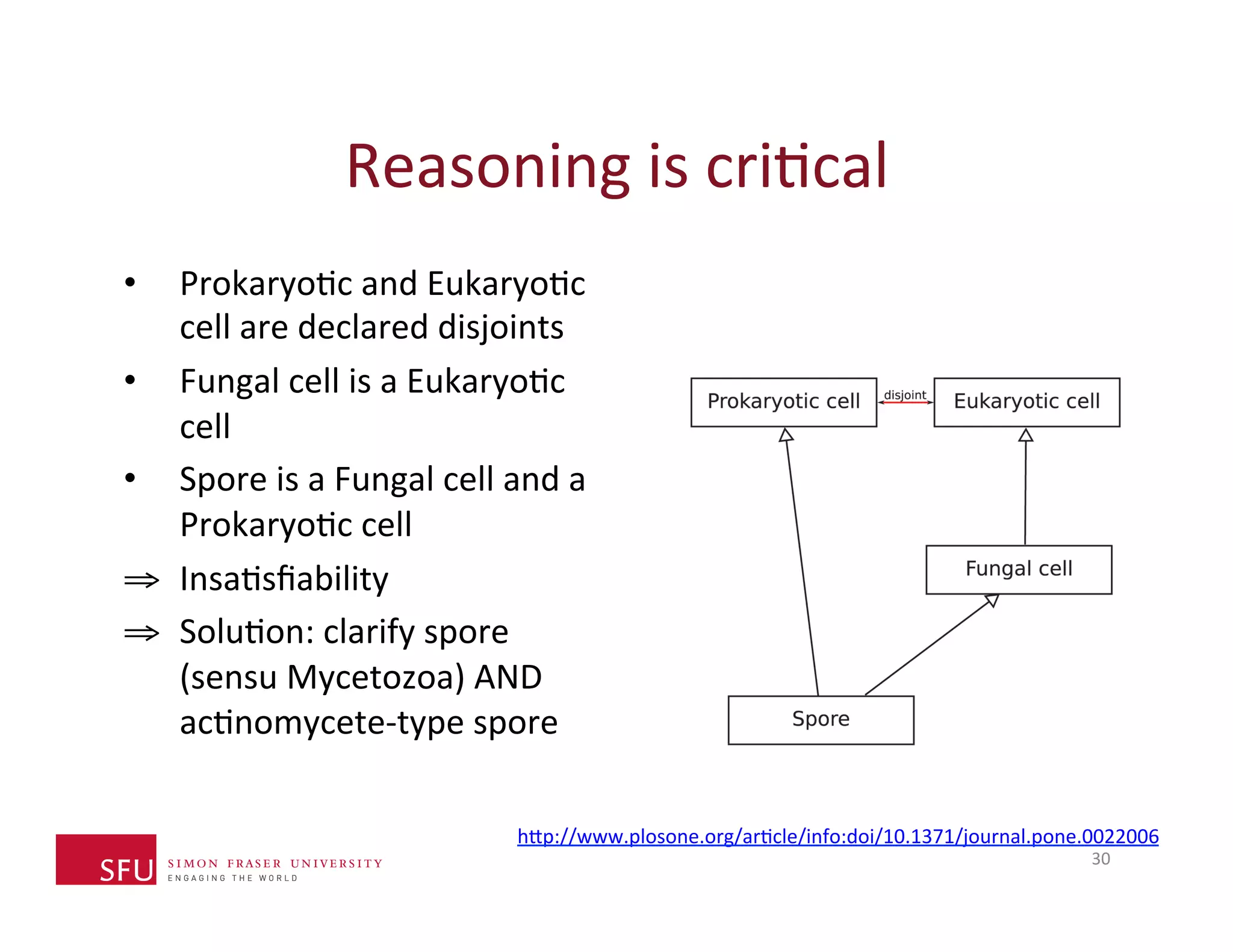
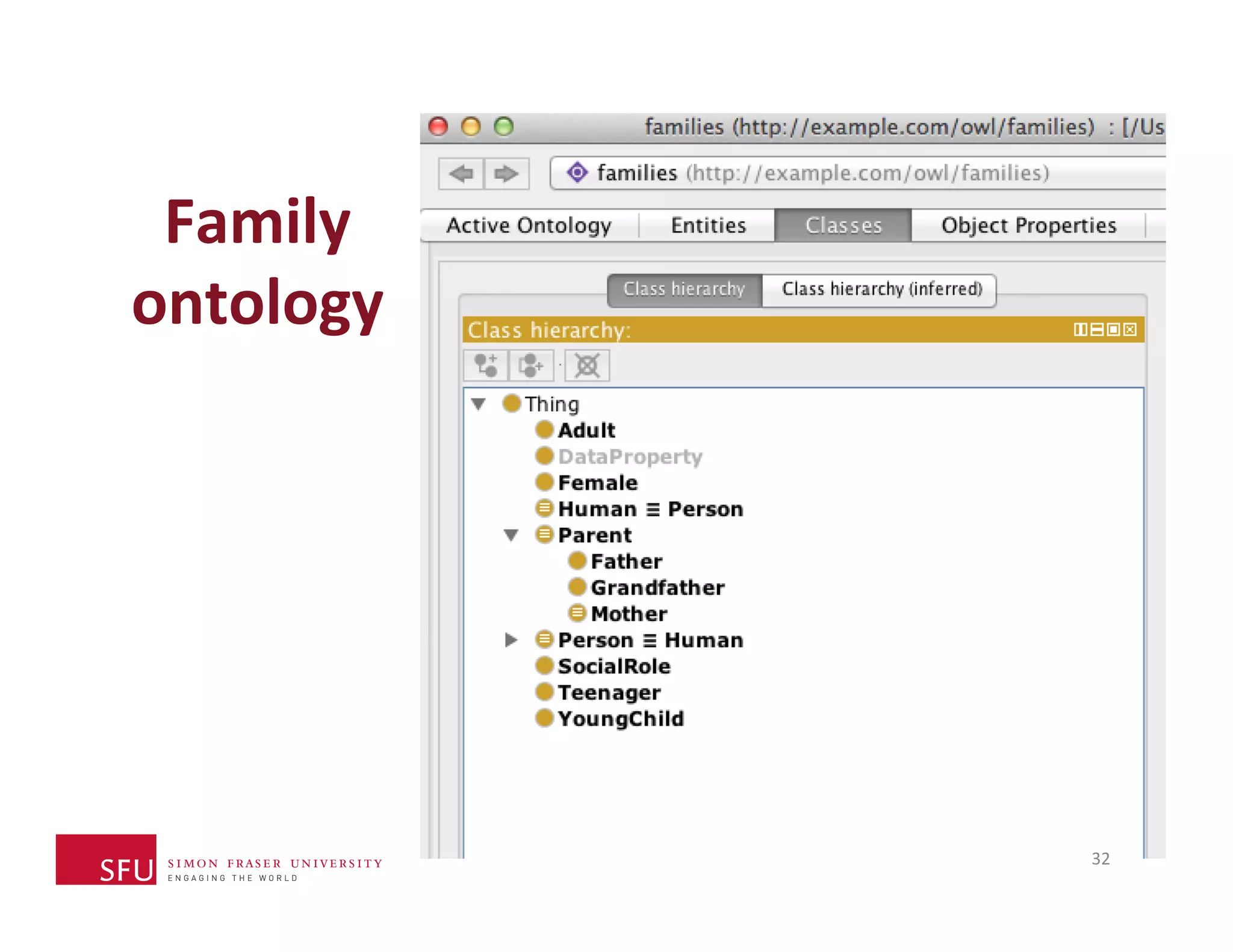
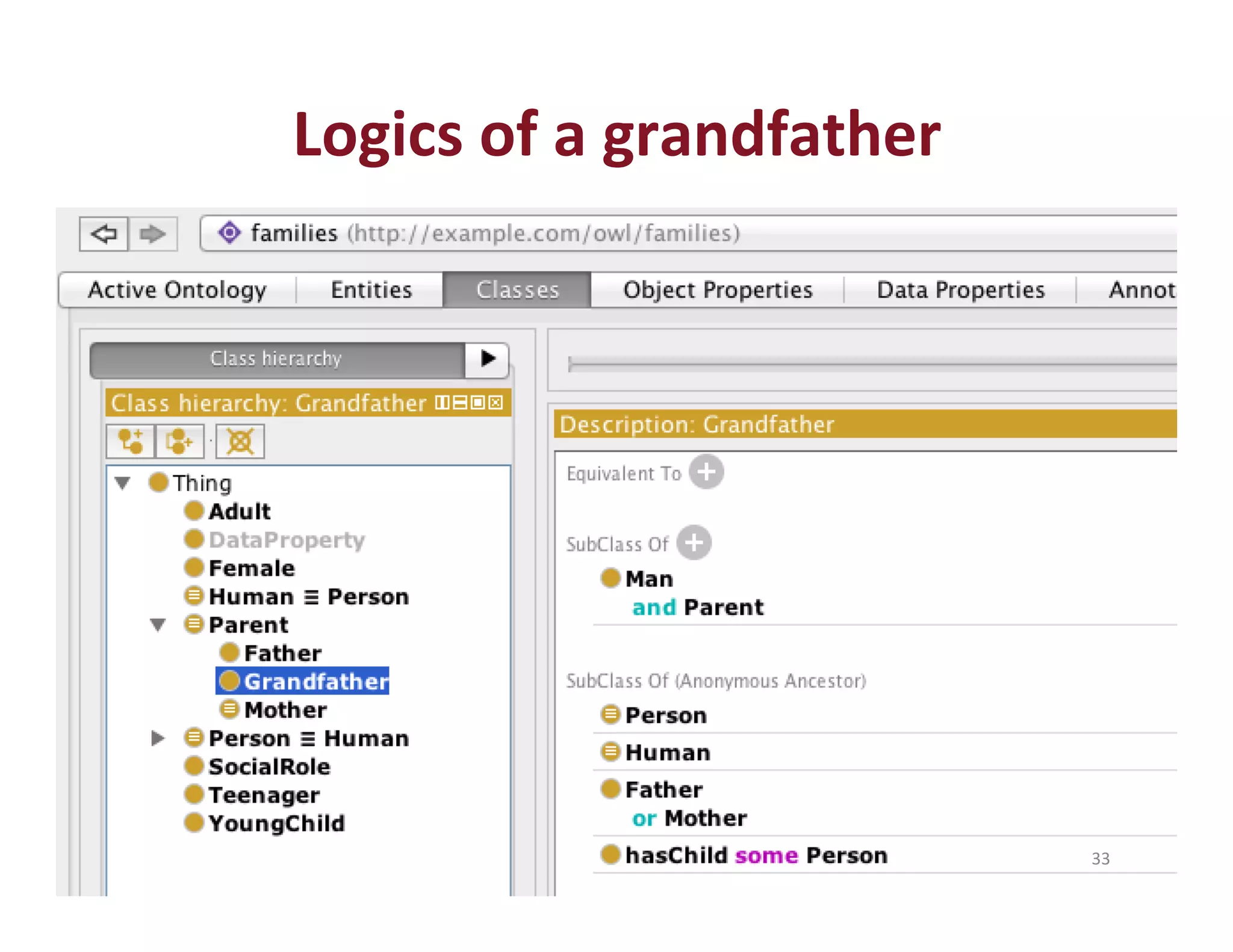
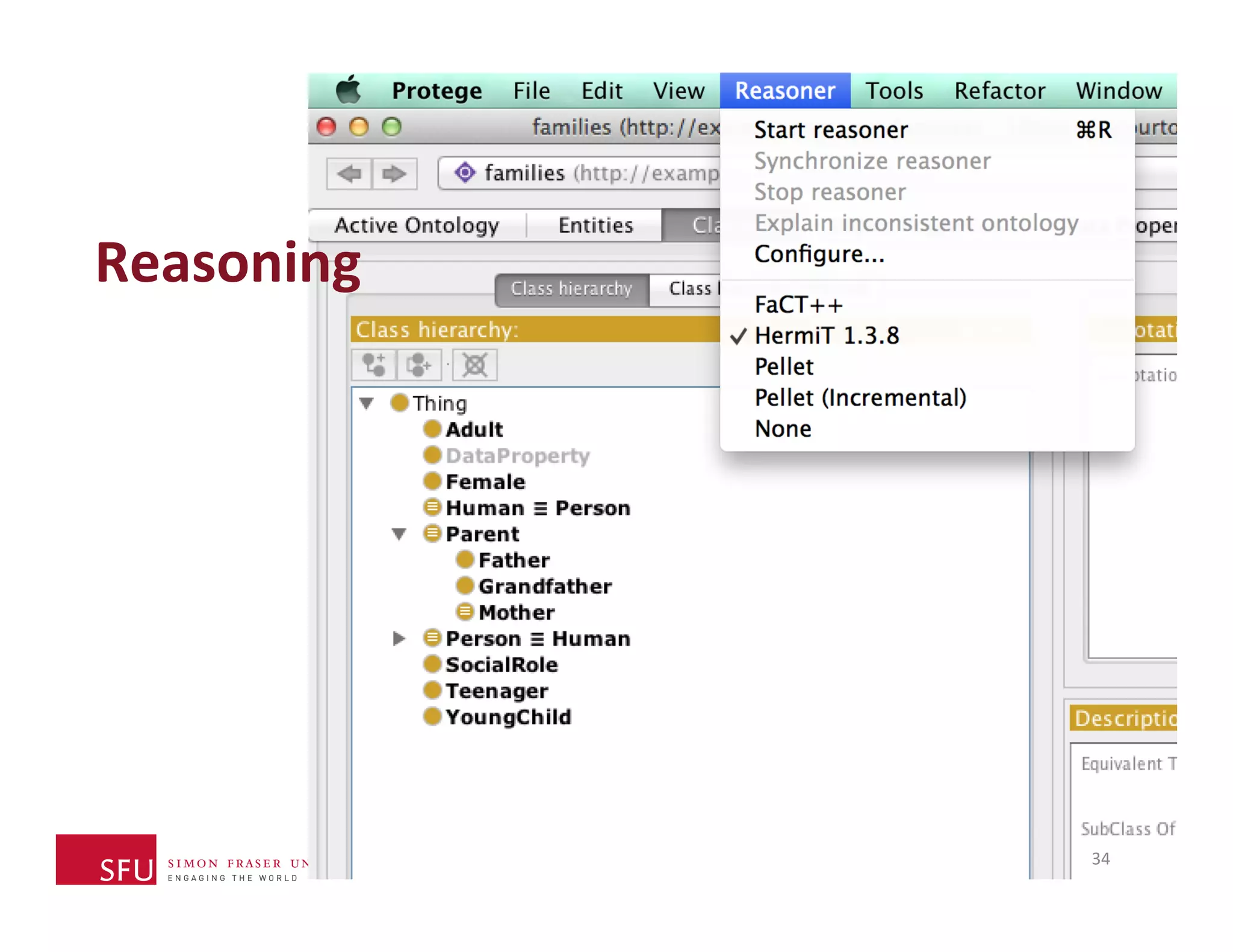
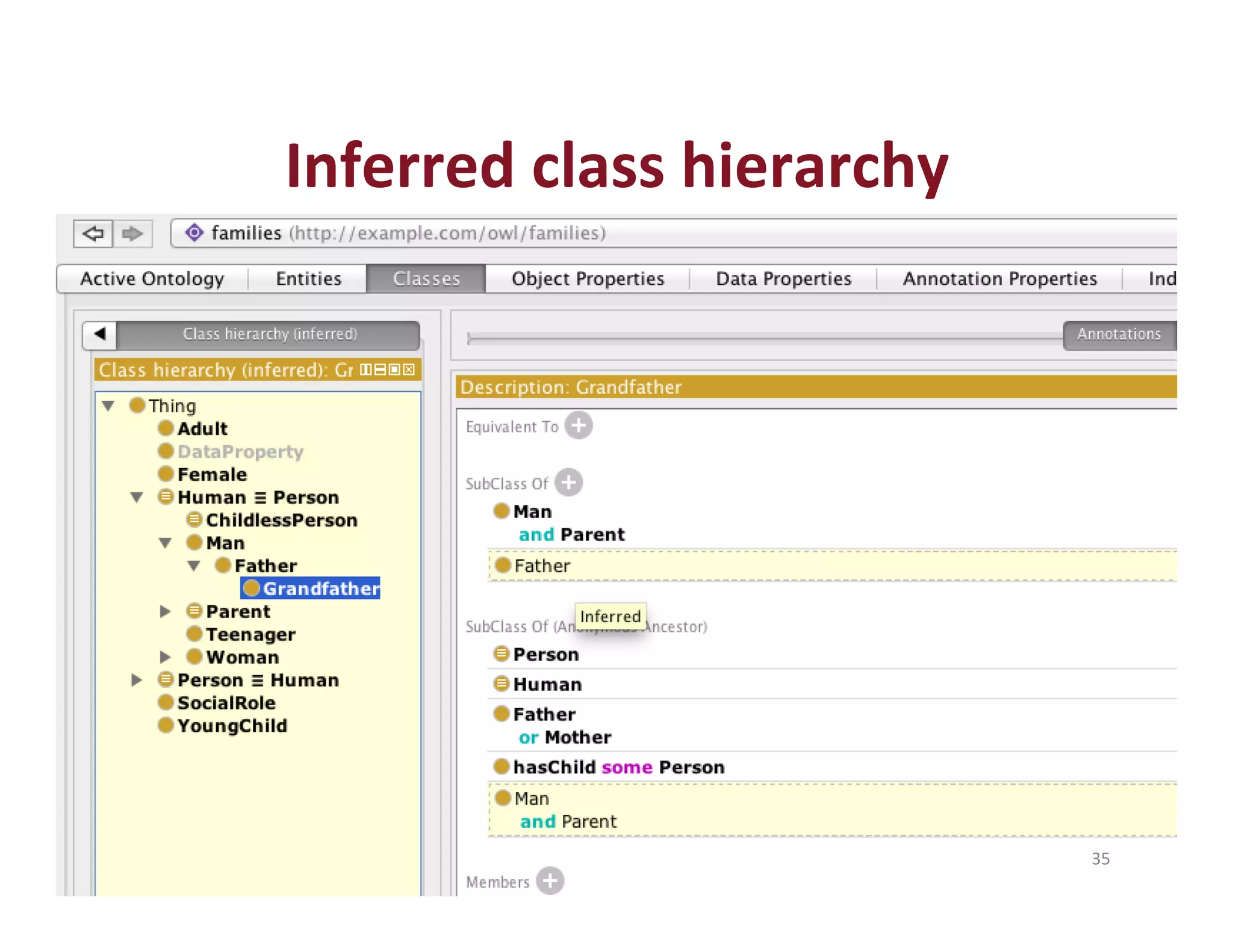
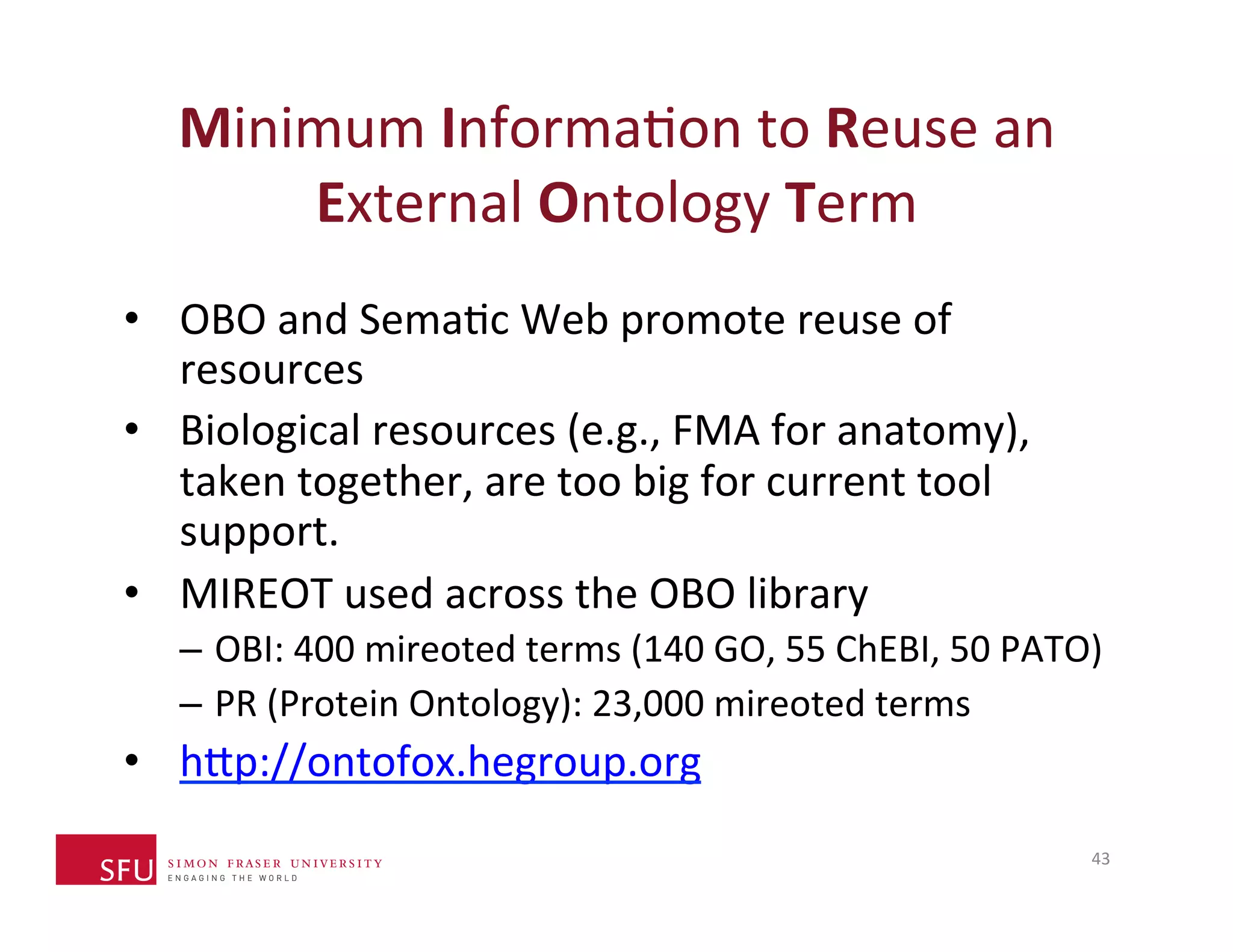
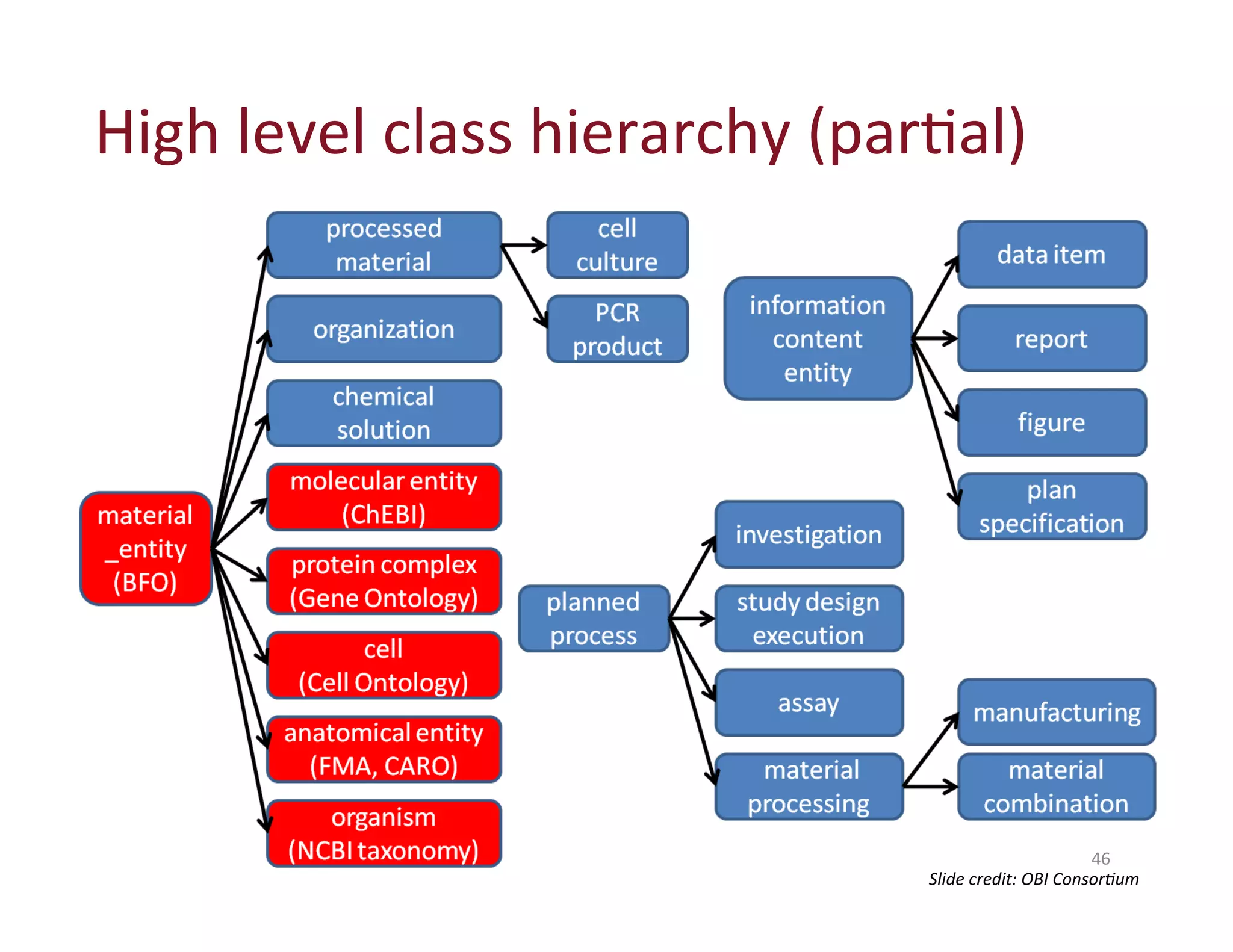
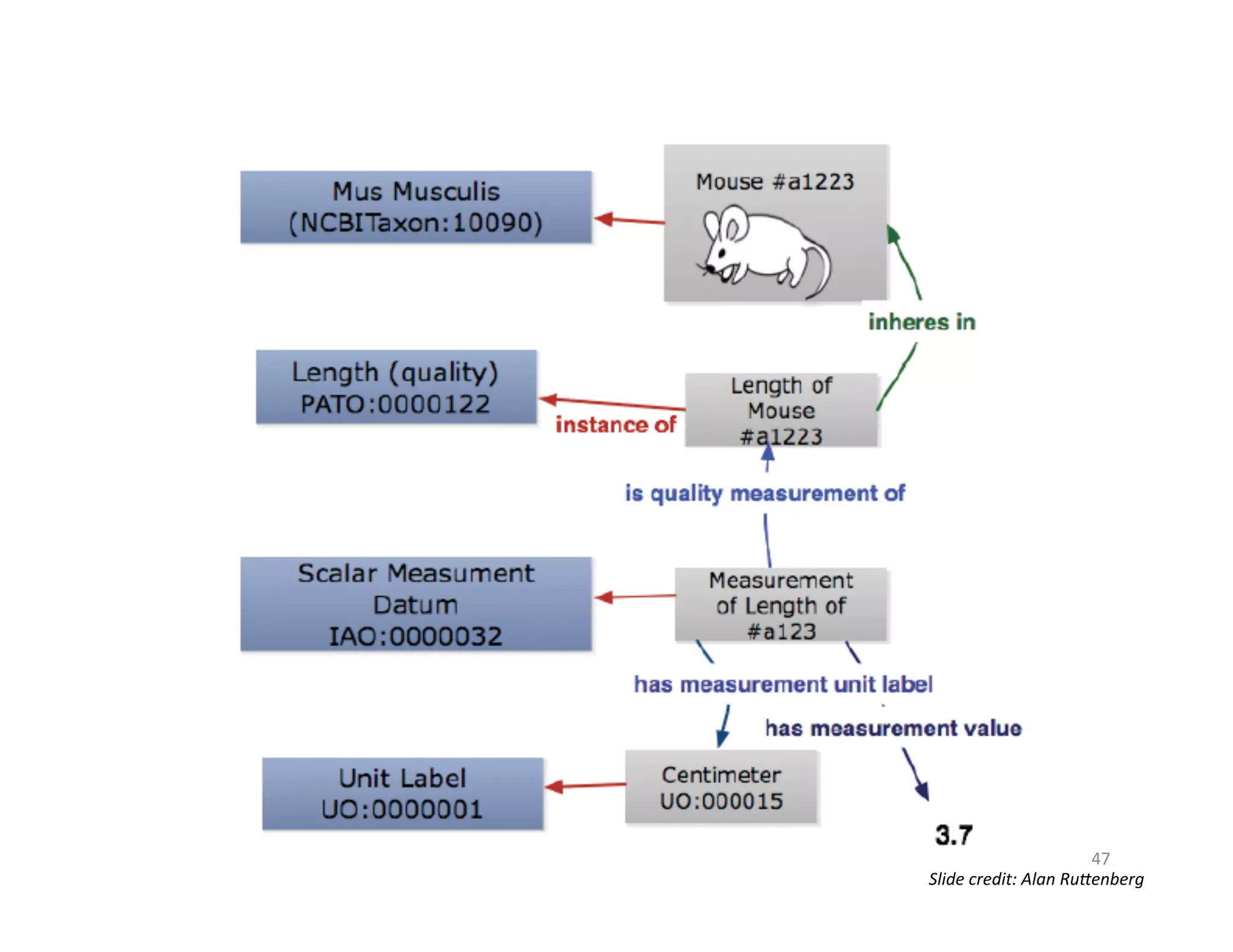
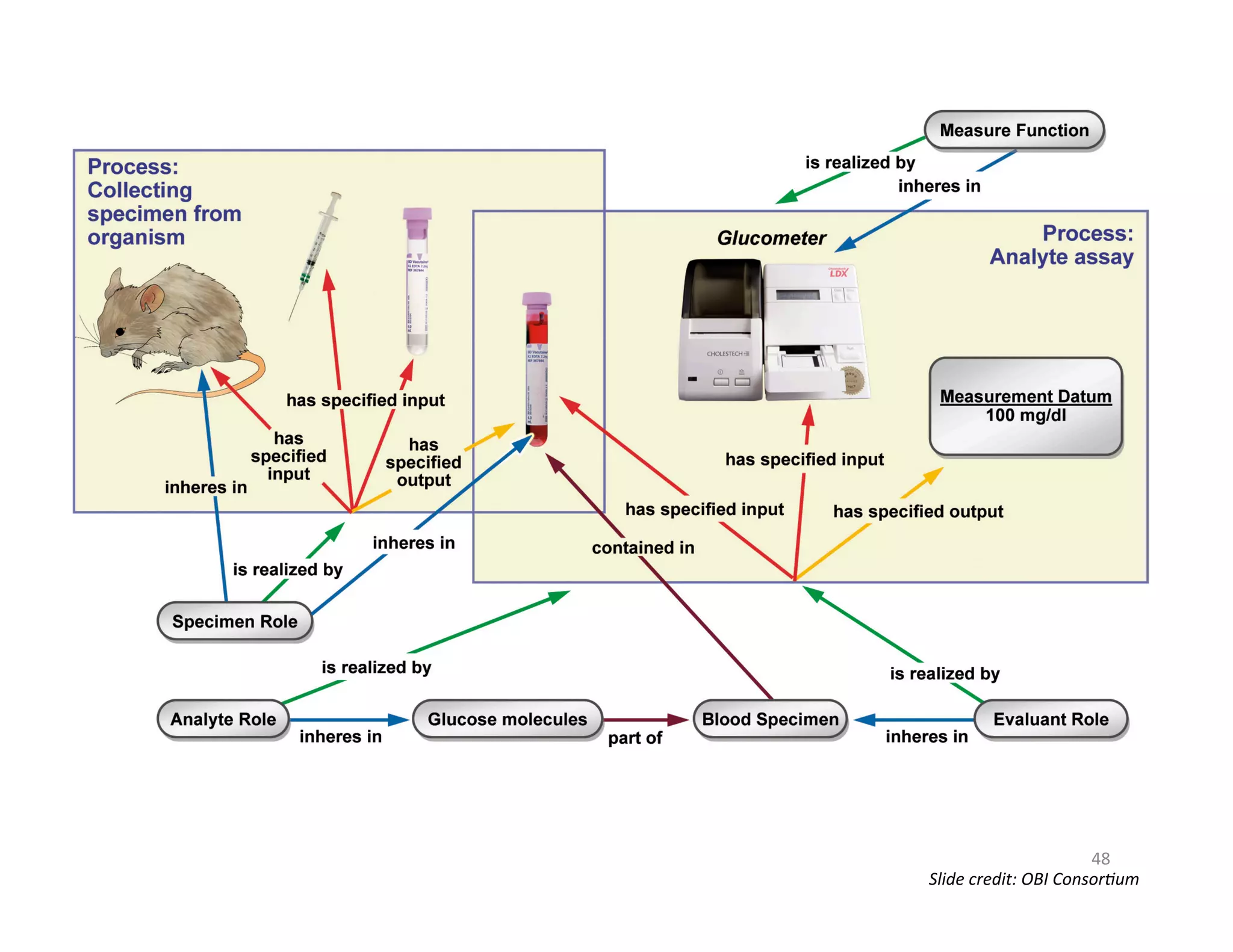
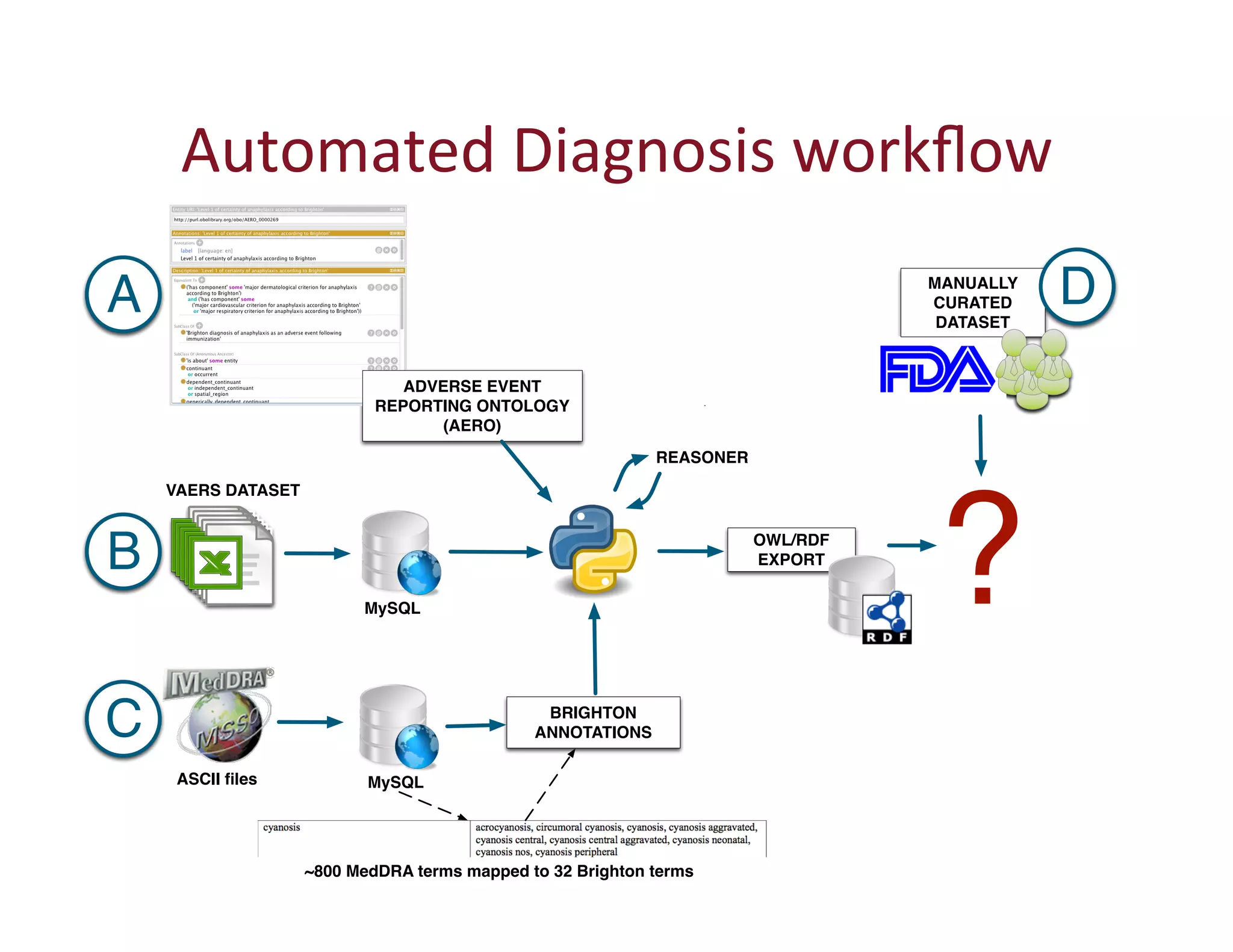
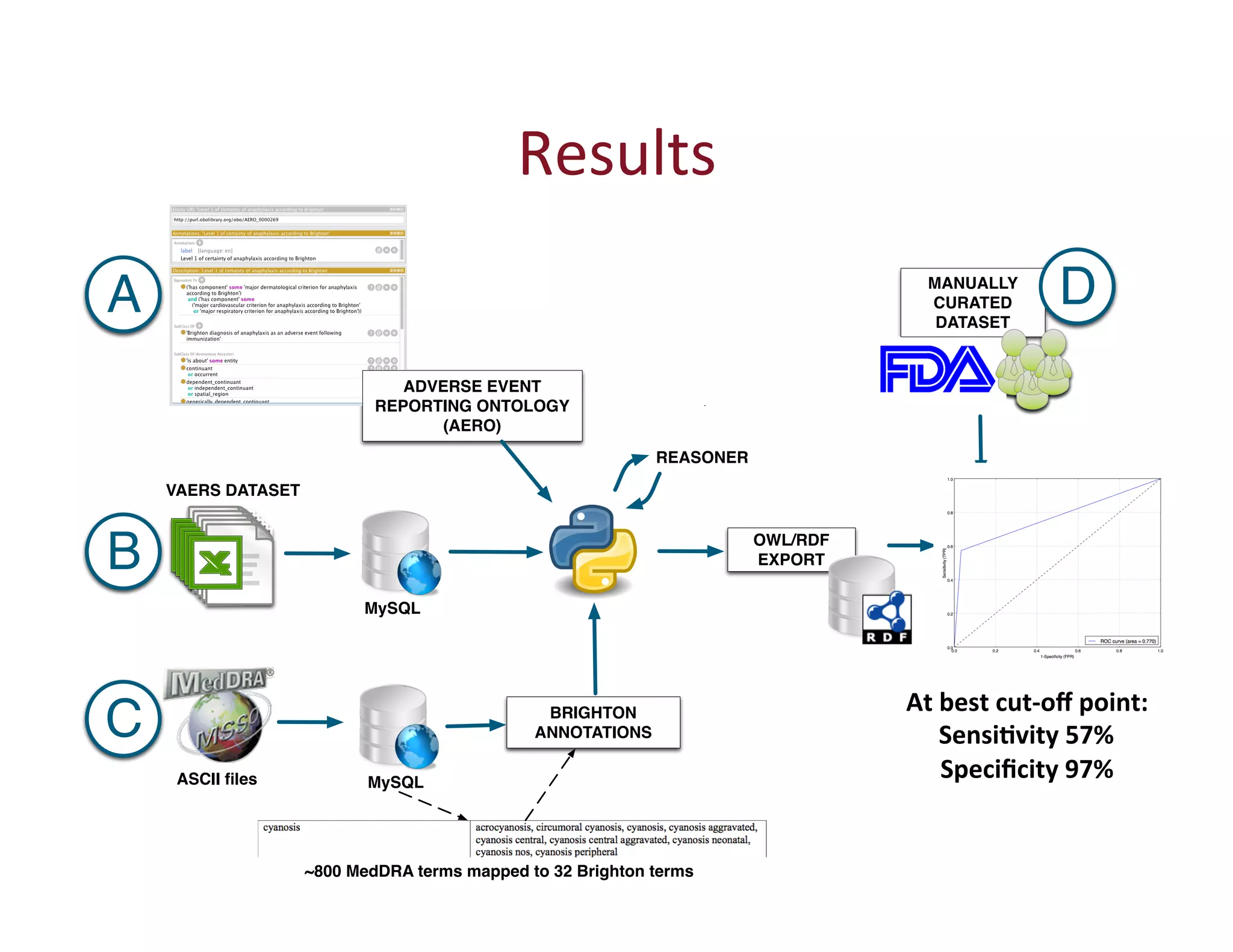
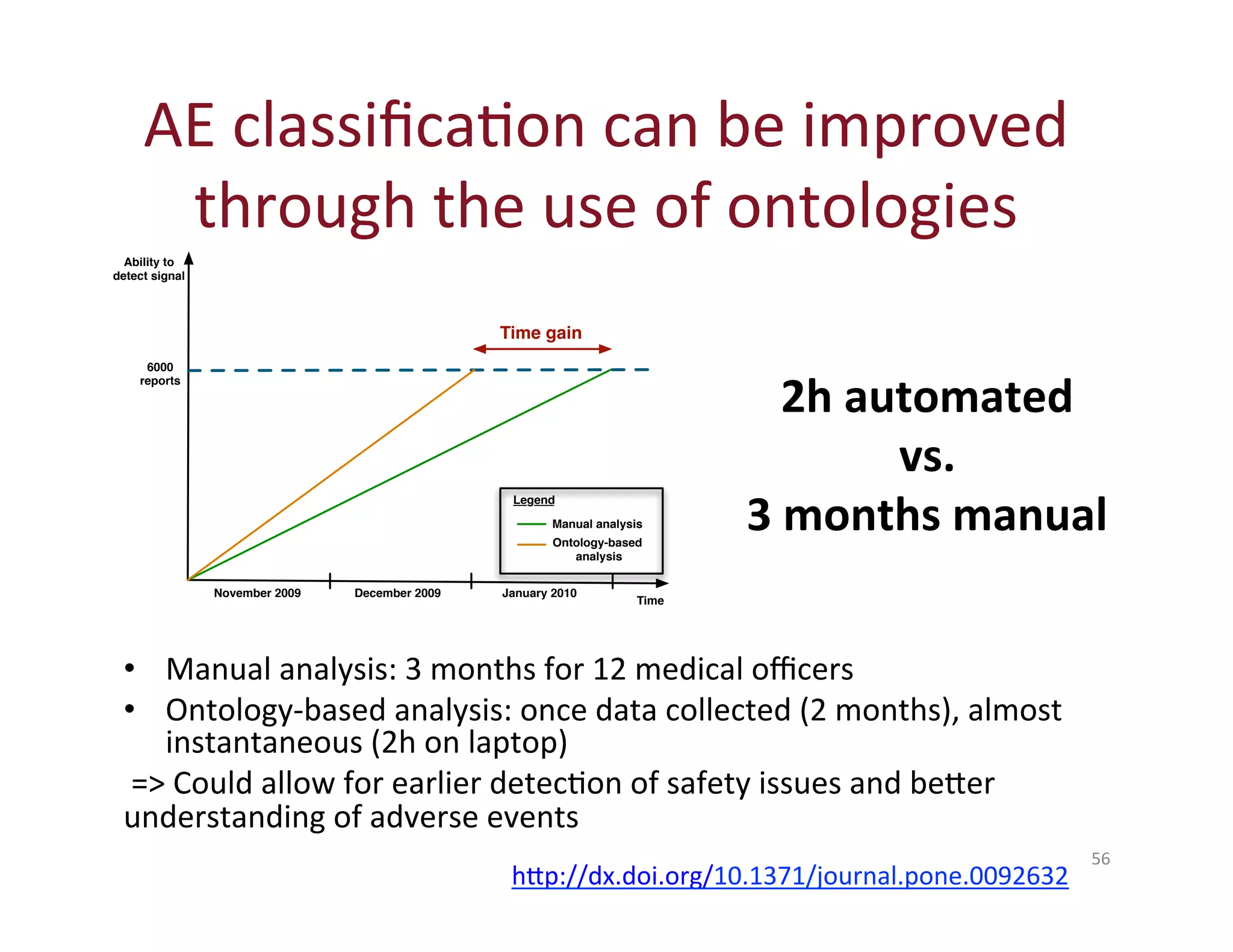
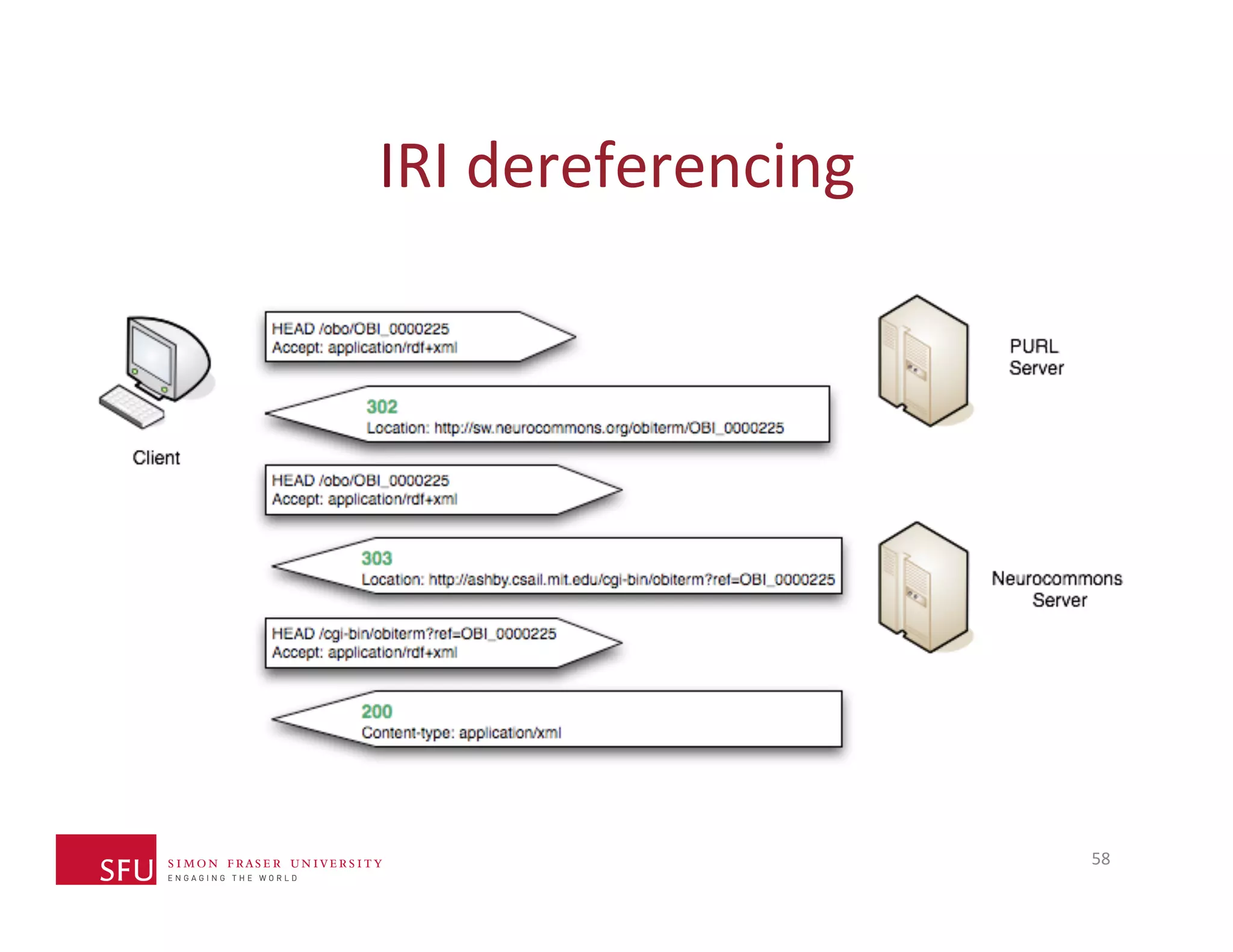
This document provides an overview of big data, the semantic web, ontologies, and the IRIDA platform. It discusses how big data is large and complex data, standards for the semantic web like URIs, RDF, SPARQL and OWL, using ontologies for definitions and reasoning, examples of existing ontologies, and how the IRIDA platform adds semantic web standards. The overview aims to provide background on these topics for analysis and integration of large and complex data sets.








![How
big
is
big?
• Facebook:
25
Terabytes
of
logged
data
per
day,
Google
(2008):
20
Petabytes
per
day
• Over
90%
of
all
the
data
in
the
world
was
created
in
the
past
2
years
[1]
• Today
3.2
ze^abytes.
2020:
40
zeAabytes.[2]
• Good
news:
jobs!
[3]
1. http://www-01.ibm.com/software/data/bigdata/
2. http://barnraisersllc.com/2012/12/38-big-facts-big-data-companies/
10
3. http://www.webopedia.com/quick_ref/important-big-data-facts-for-it-professionals.html](https://image.slidesharecdn.com/20141112courtotbigdatasemwebontologies-141117152909-conversion-gate02/75/20141112-courtot-big_datasemwebontologies-10-2048.jpg)