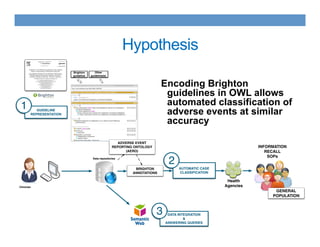
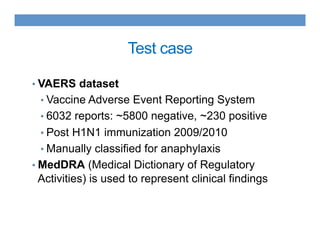
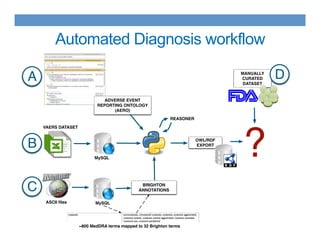
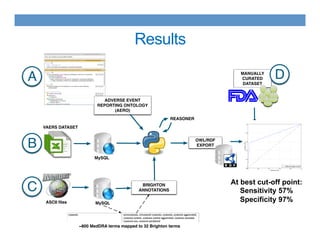
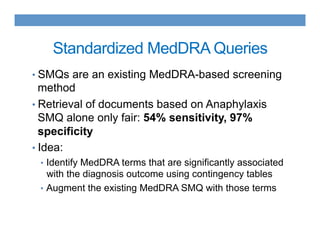
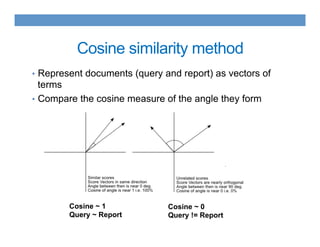
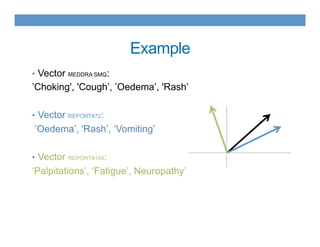
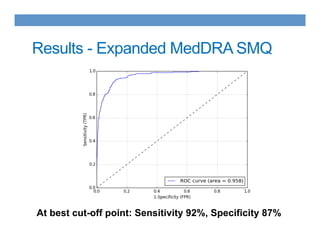
This document discusses using an ontology-based approach to automatically classify adverse event reports at a similar accuracy as manual classification. It tested classifying over 6000 vaccine adverse event reports for anaphylaxis using terms from an adverse event reporting ontology mapped to guidelines. The automated approach achieved a maximum sensitivity of 57% and specificity of 97%. Additional techniques improved sensitivity to 92% while maintaining high specificity. The results demonstrate the potential for ontologies to help analyze large datasets of adverse event reports.