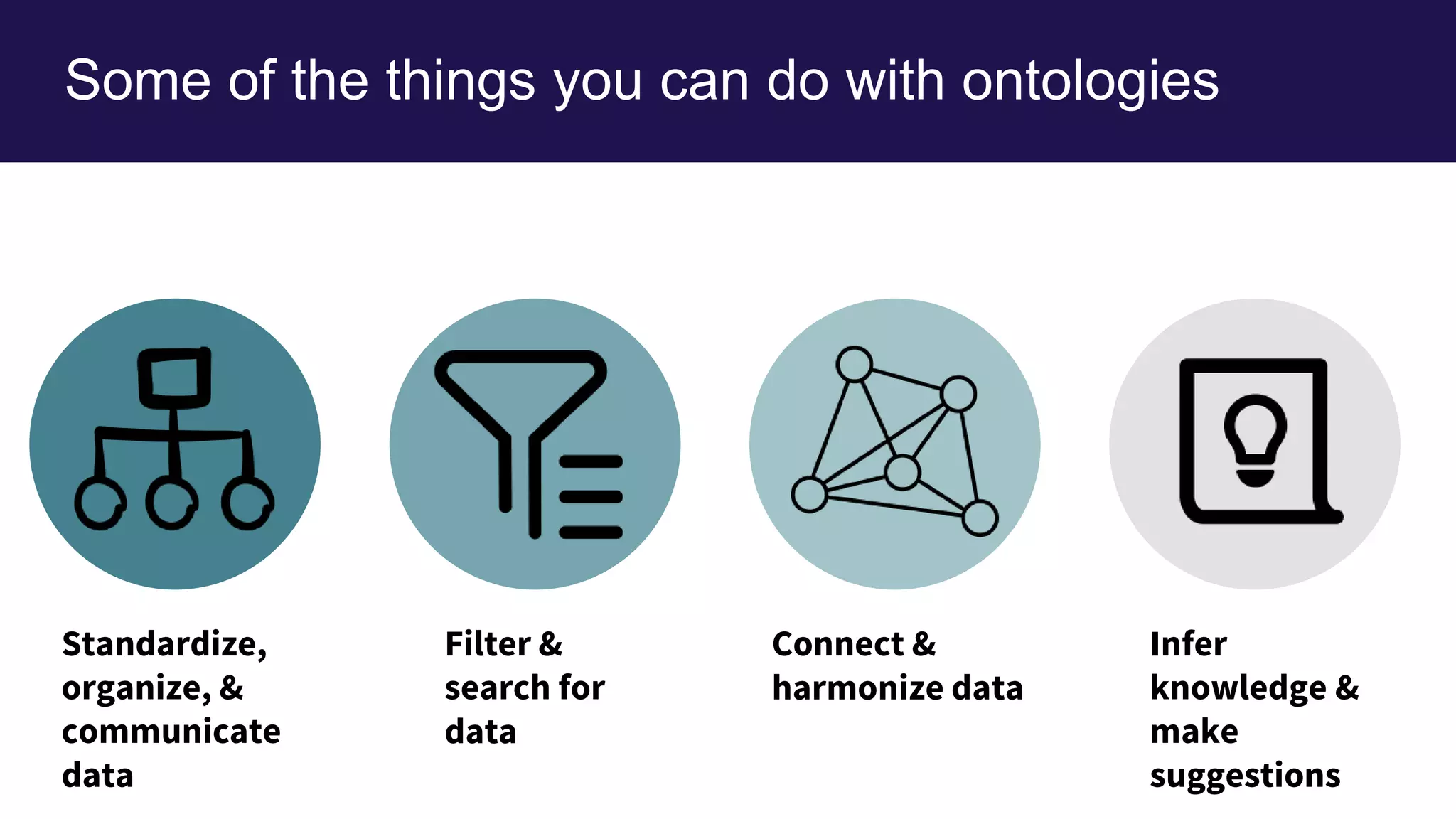
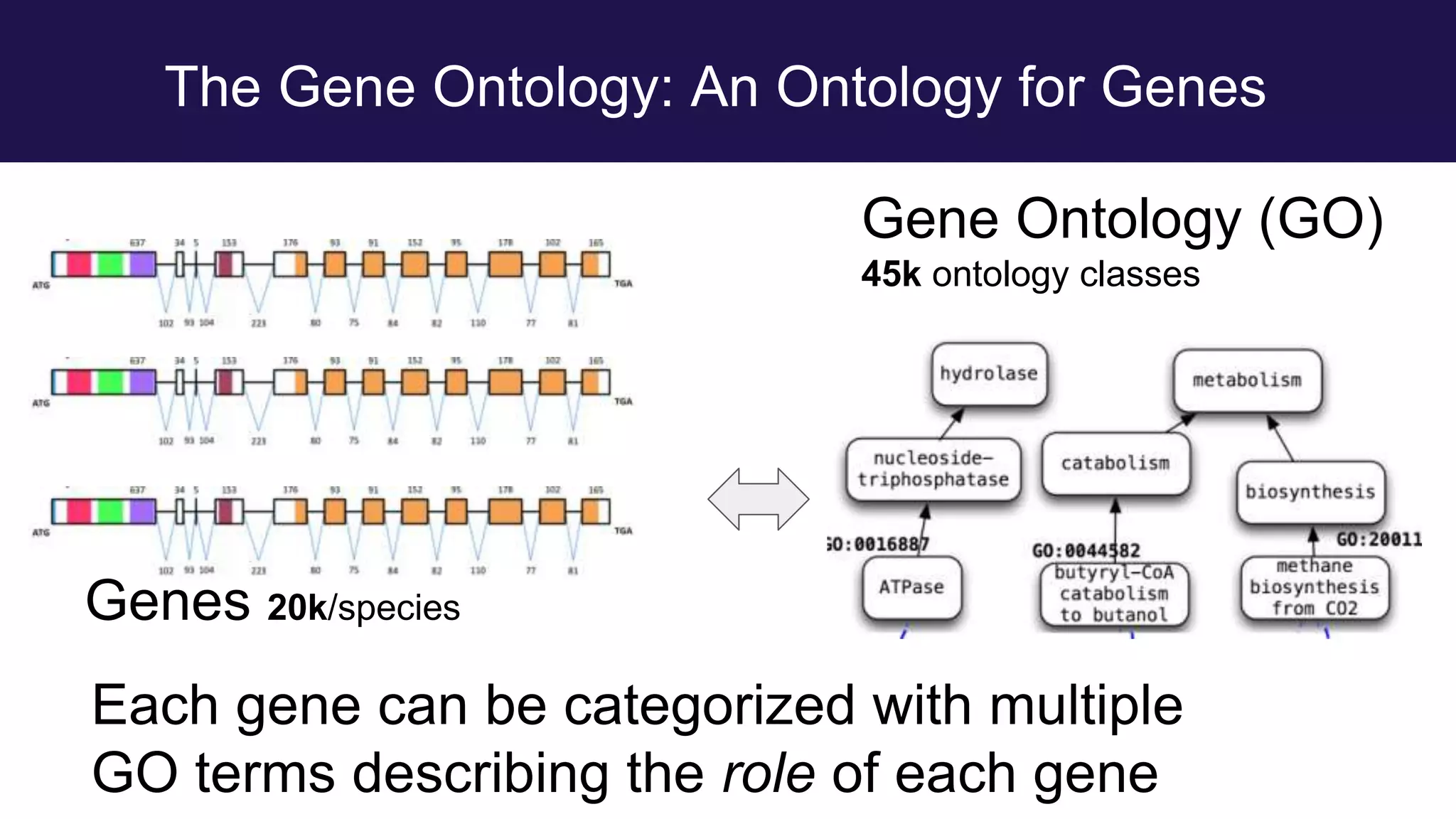
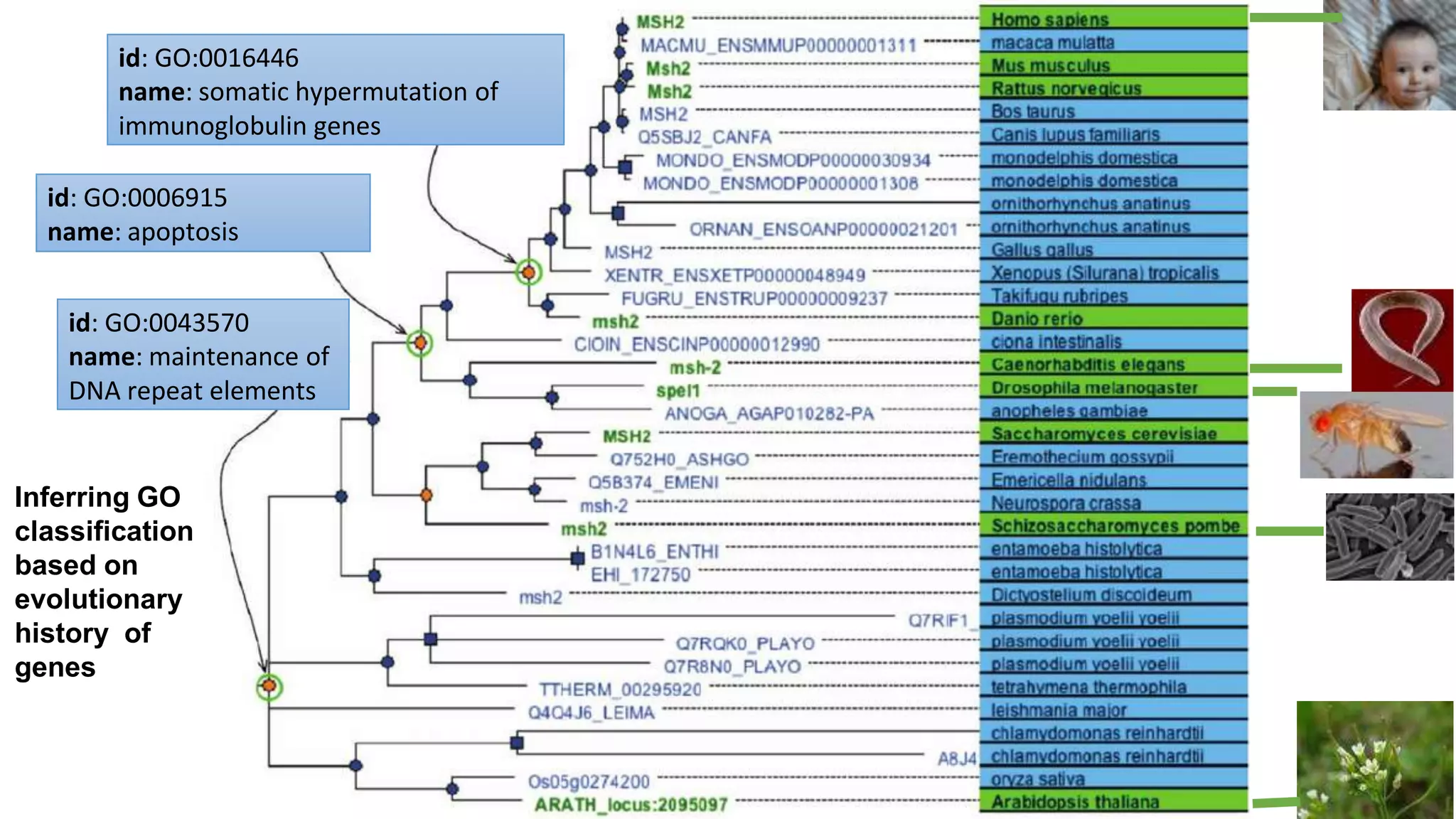
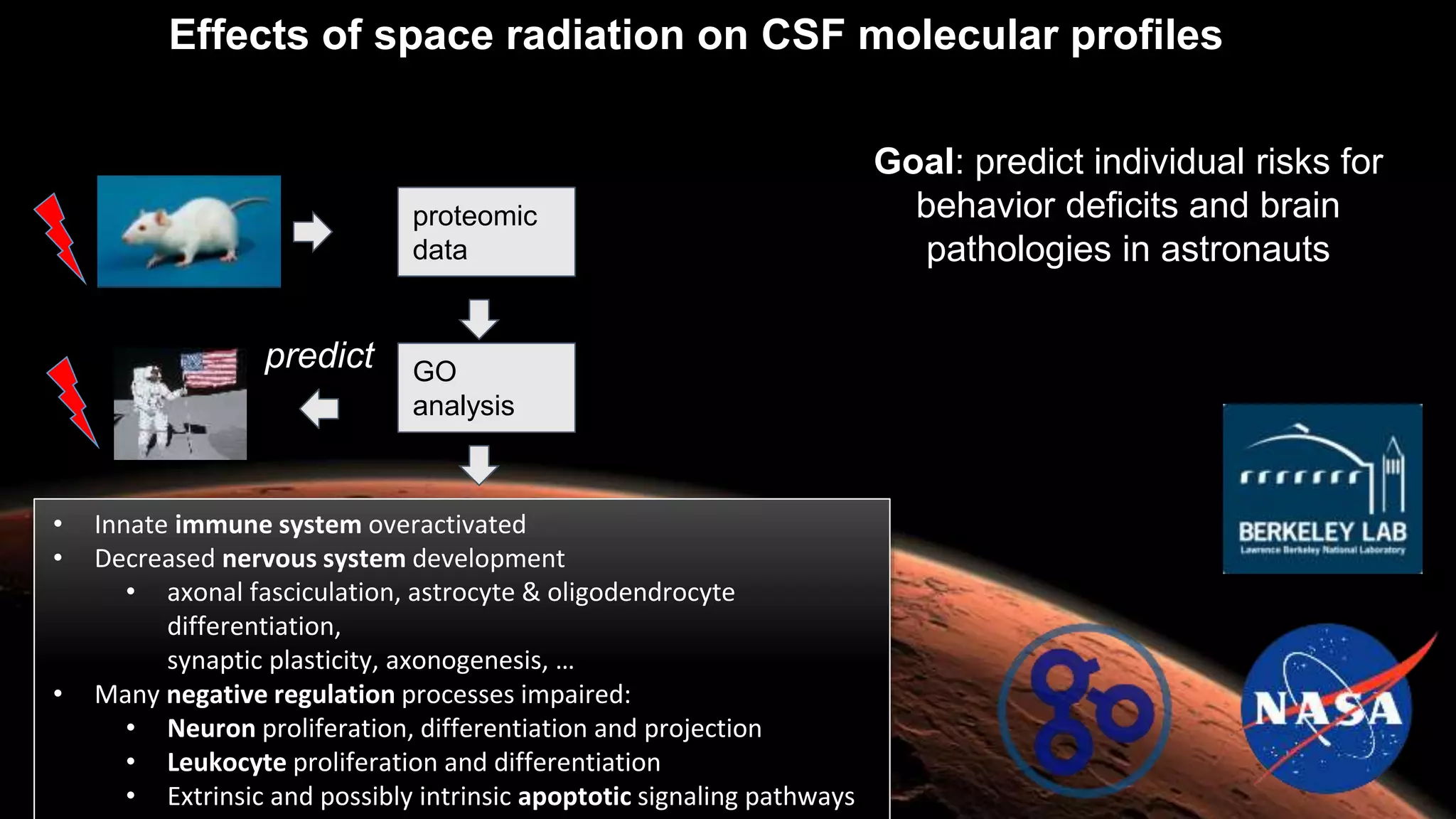
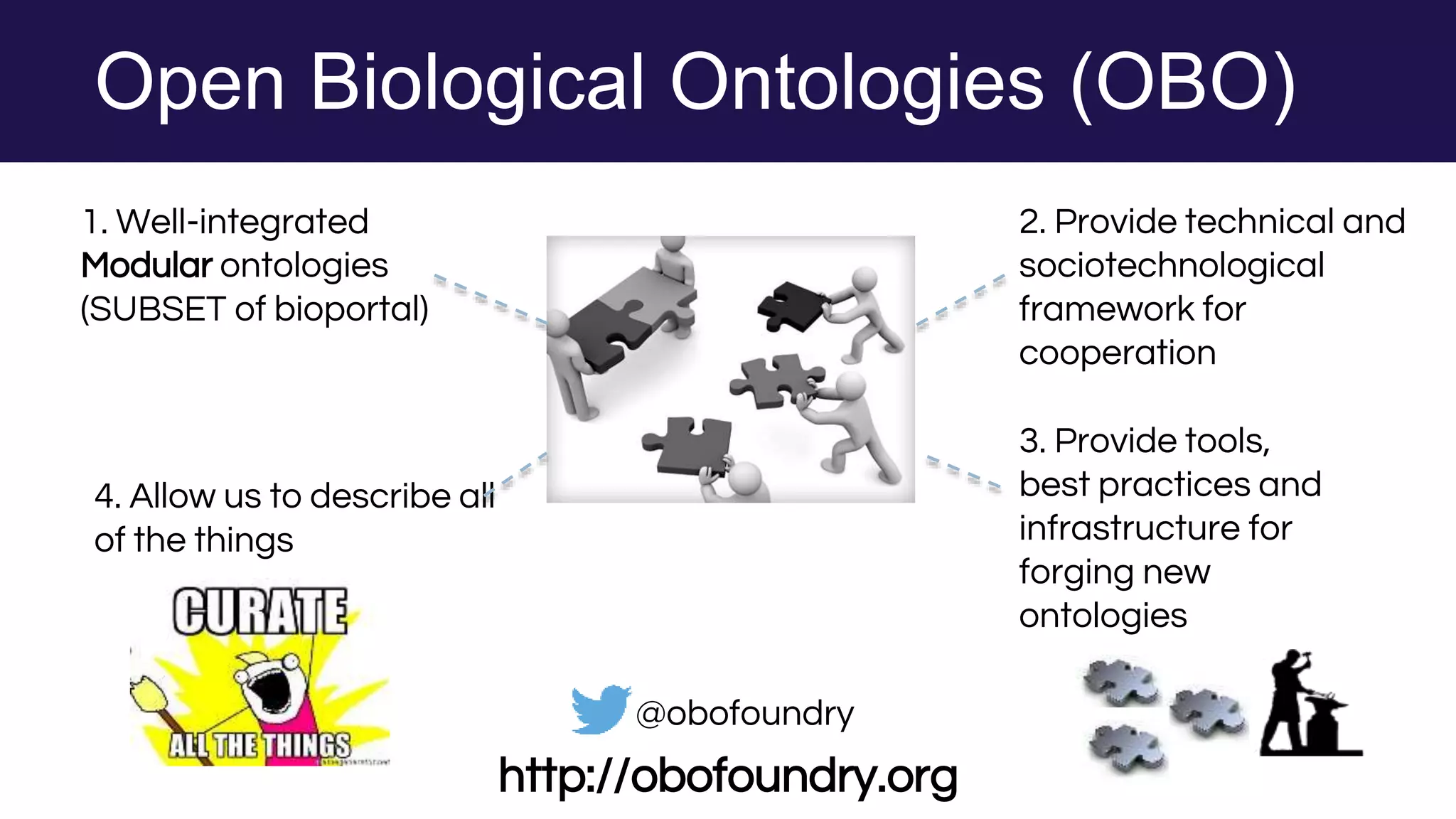
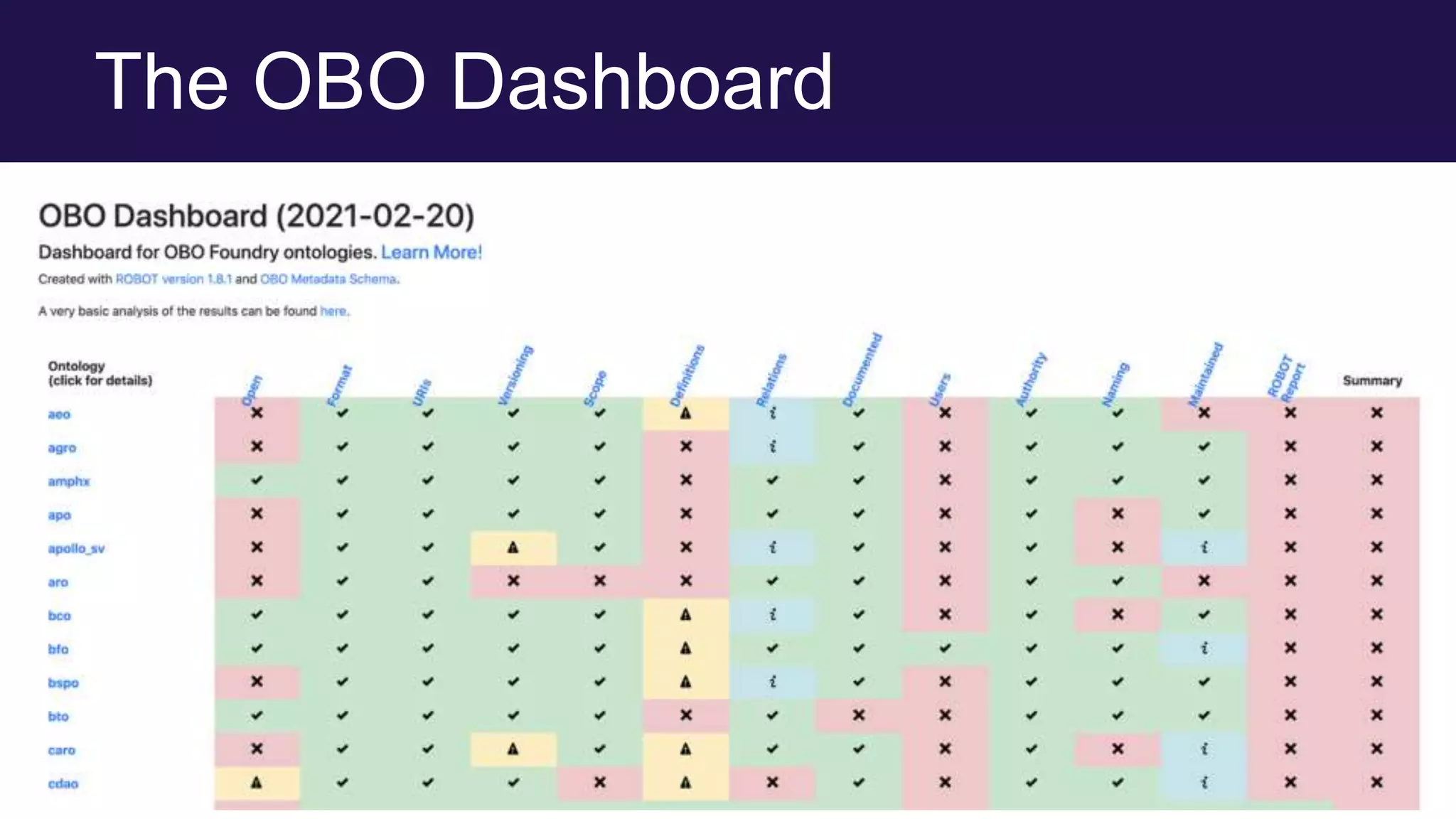
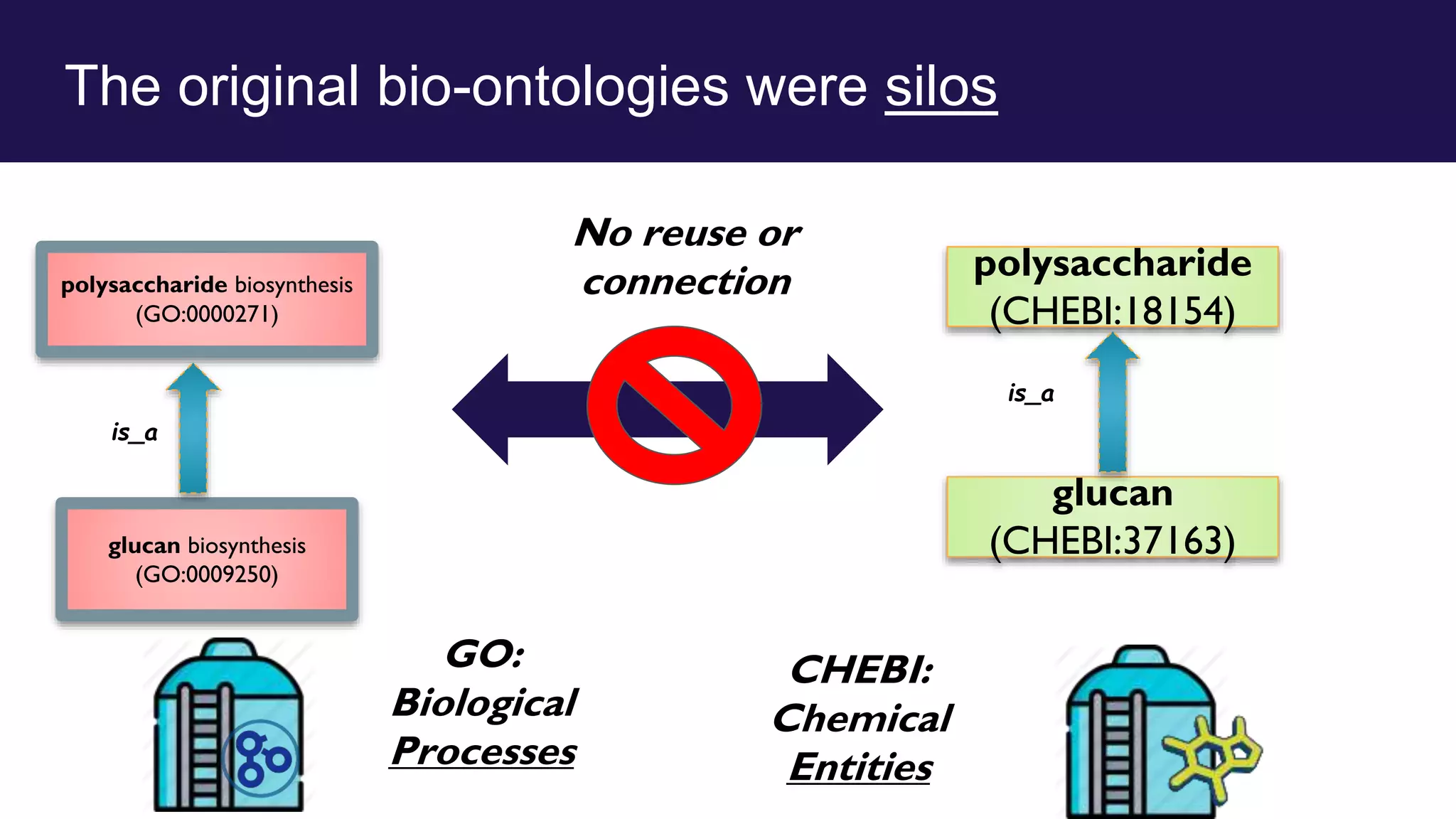
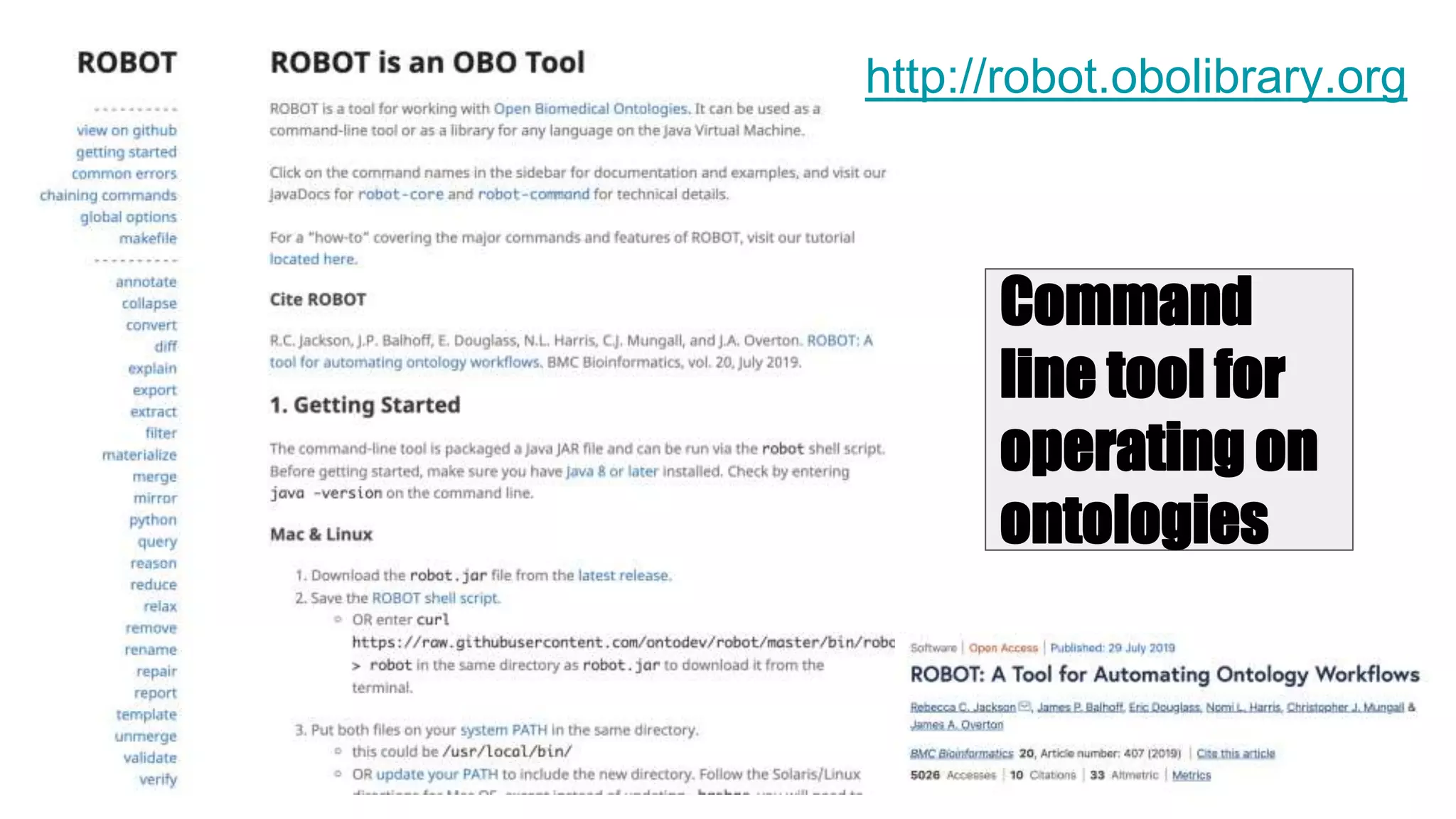
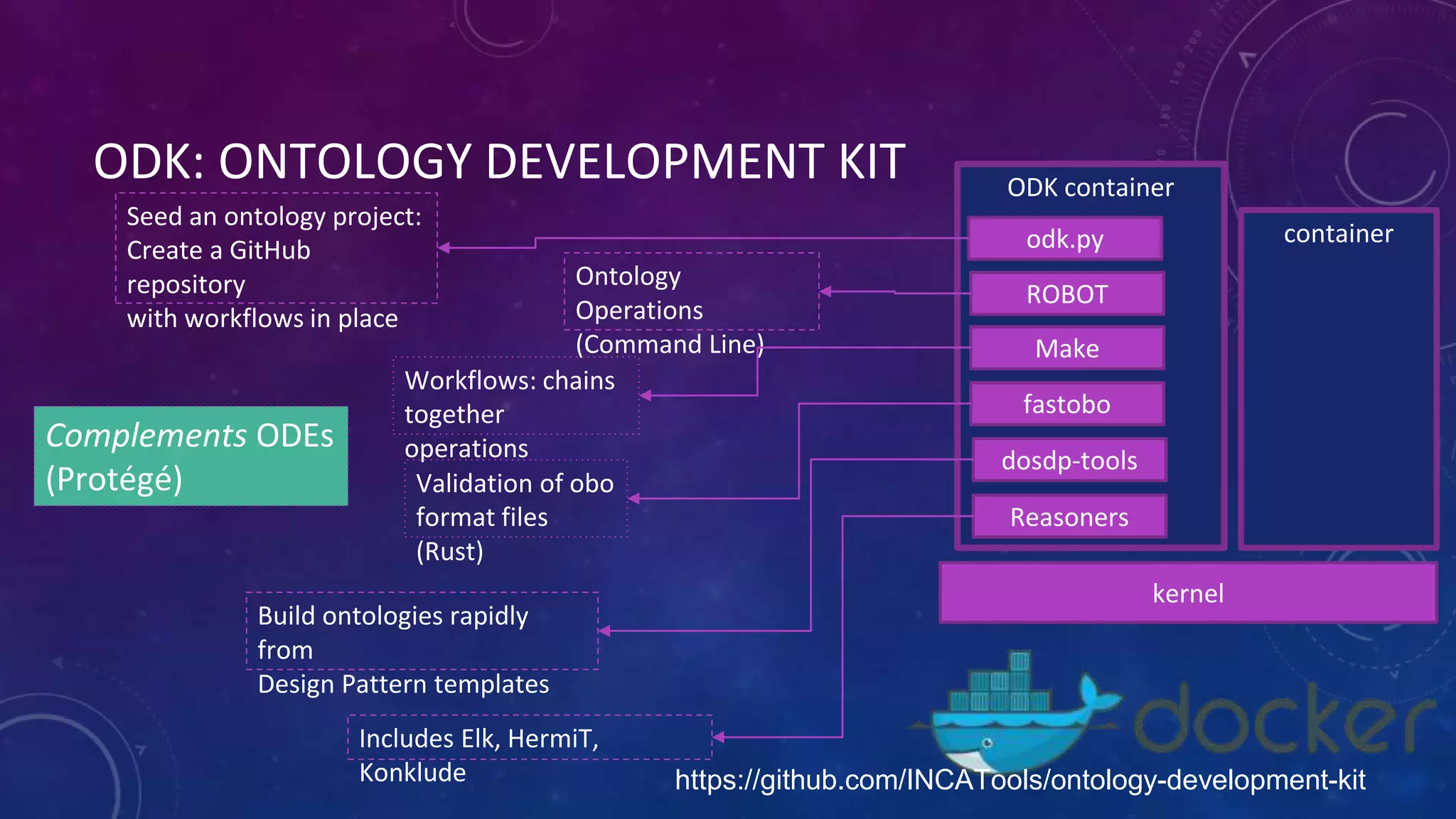
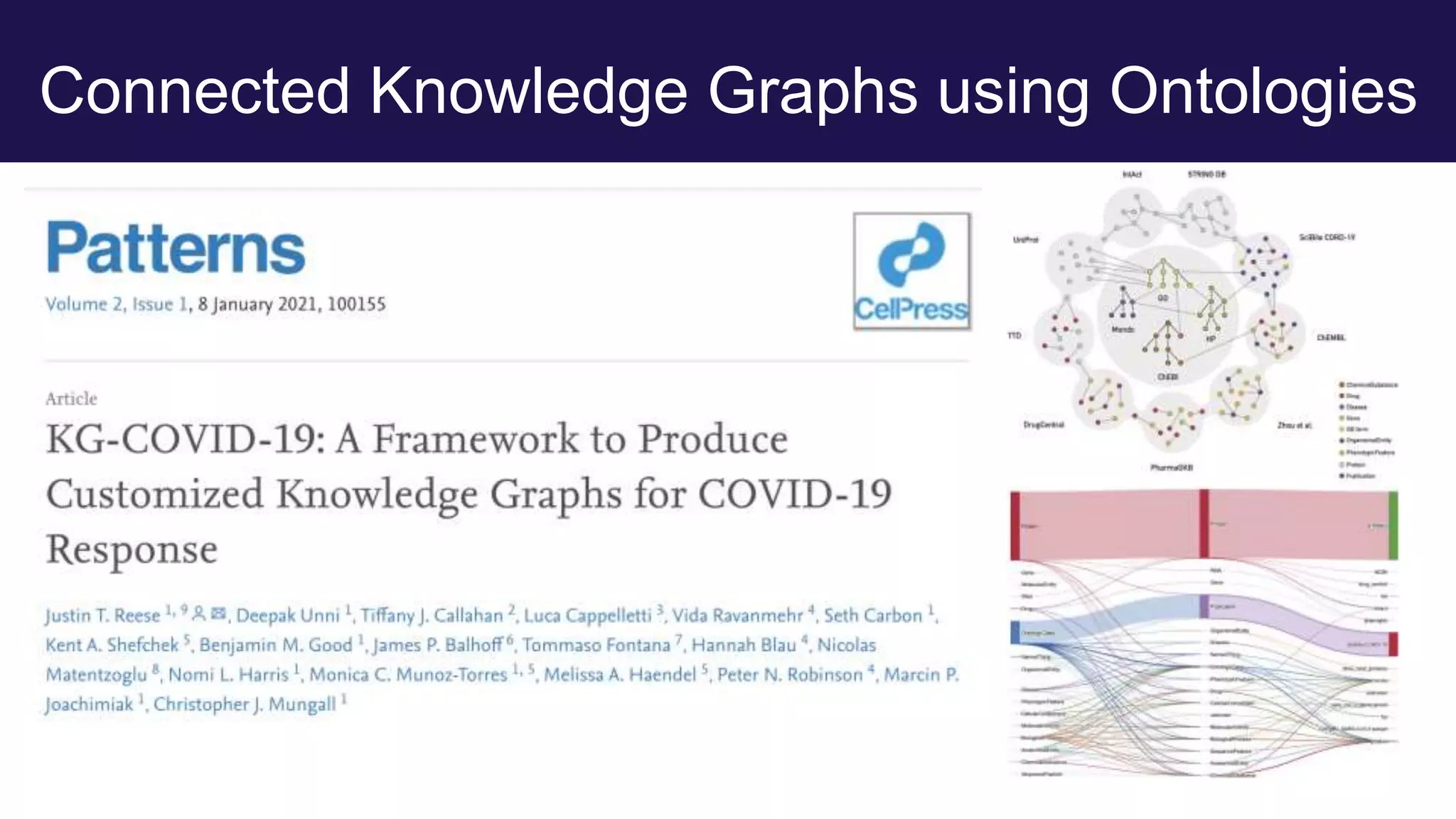
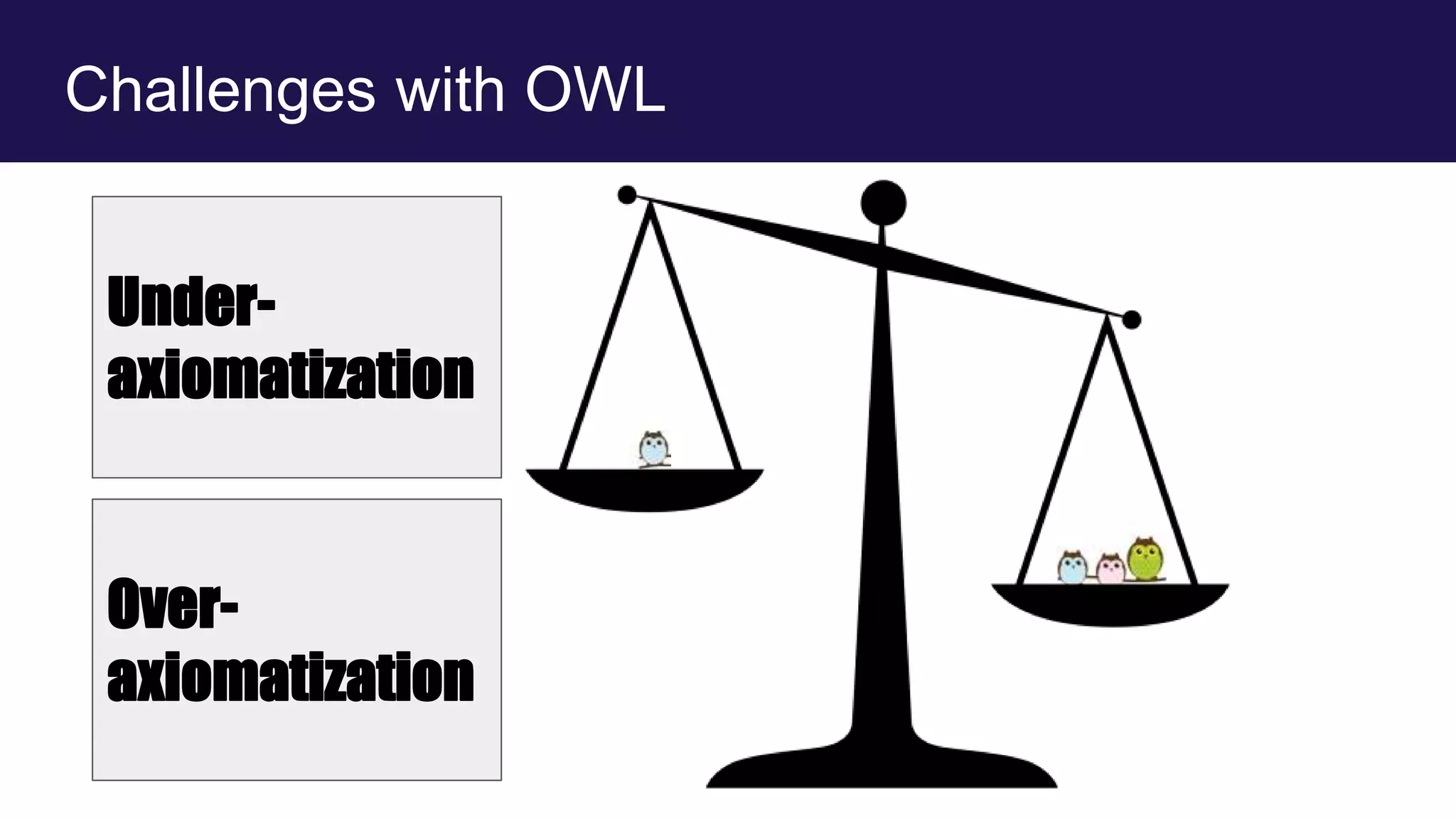
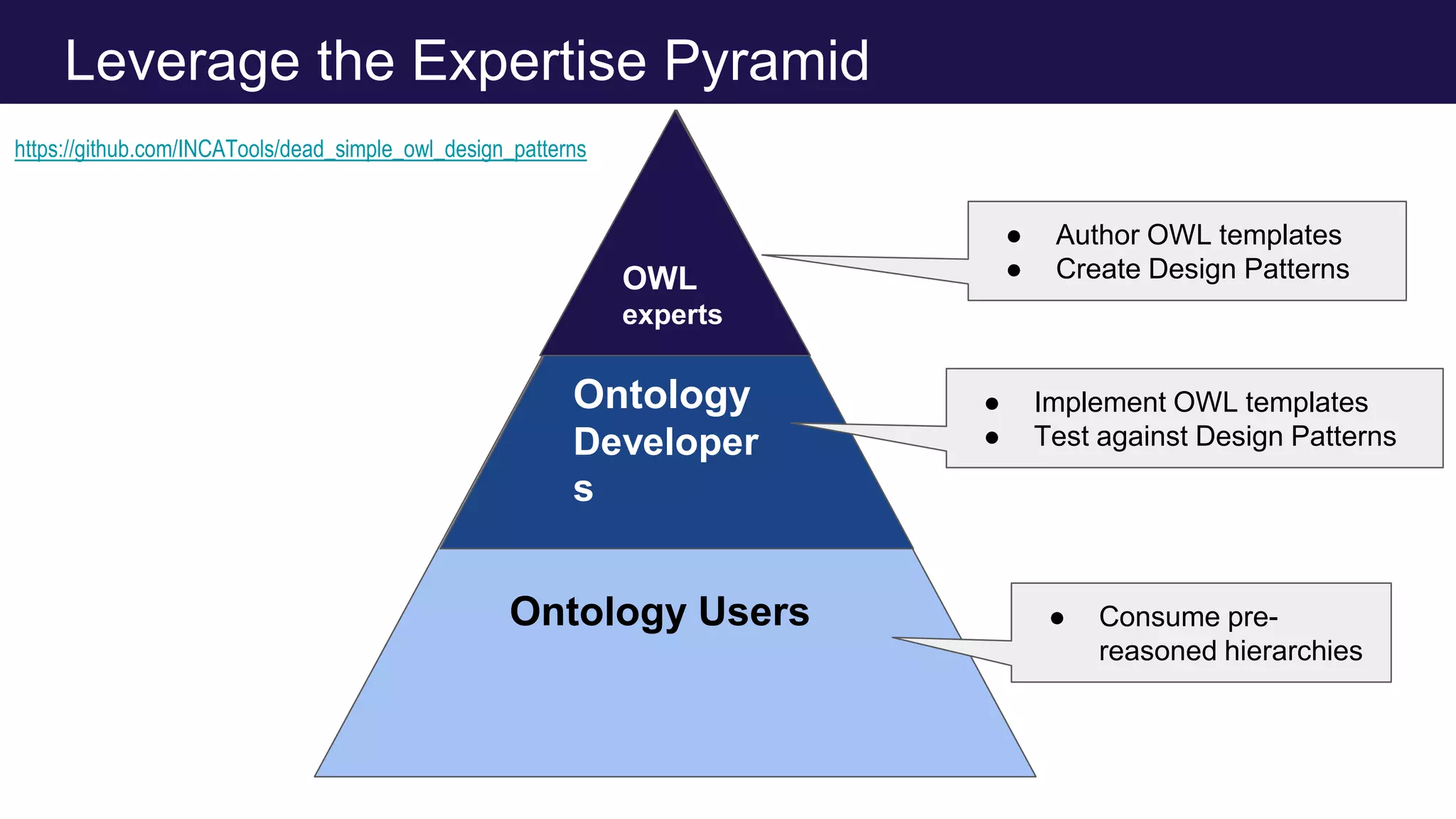
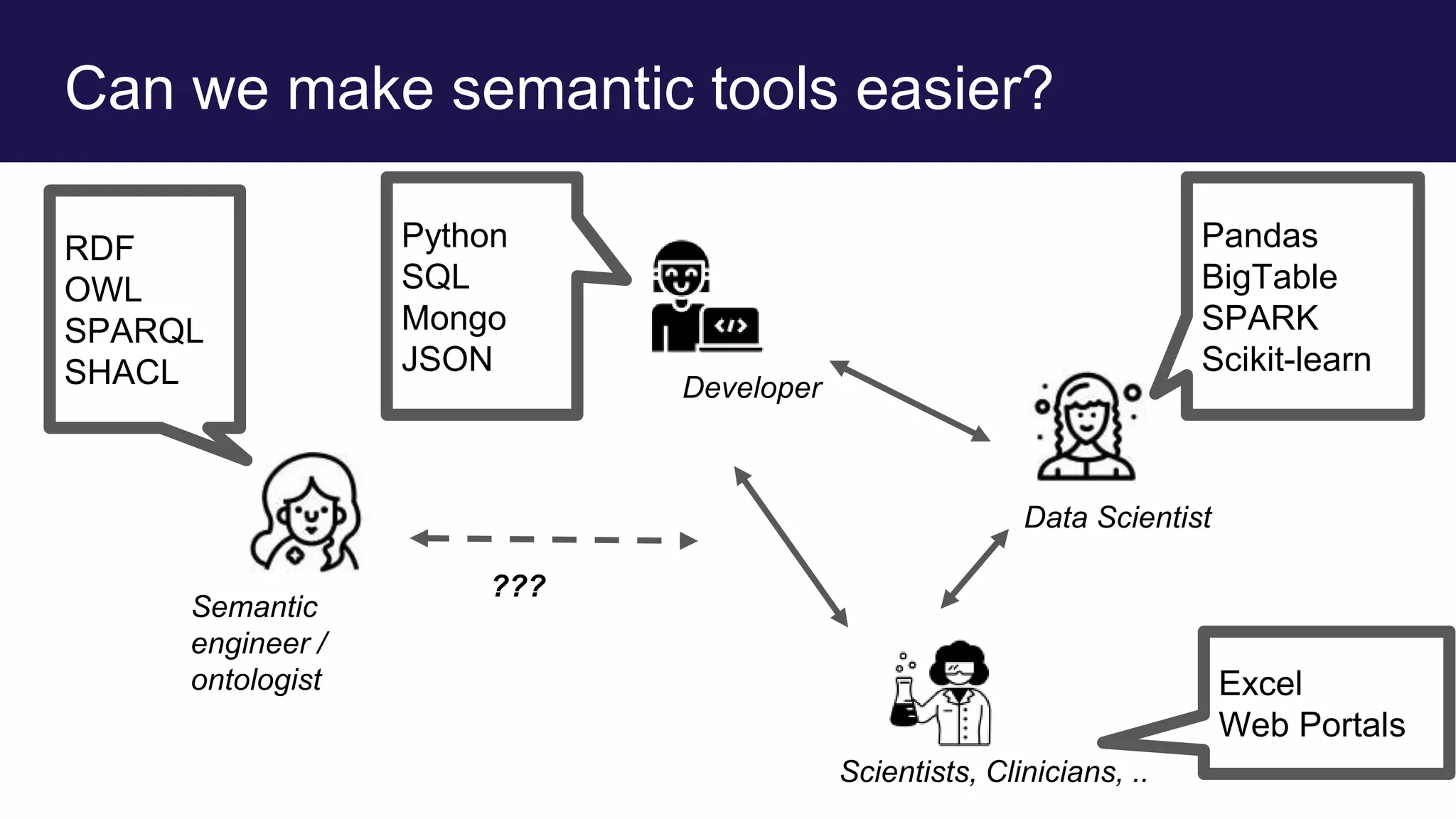
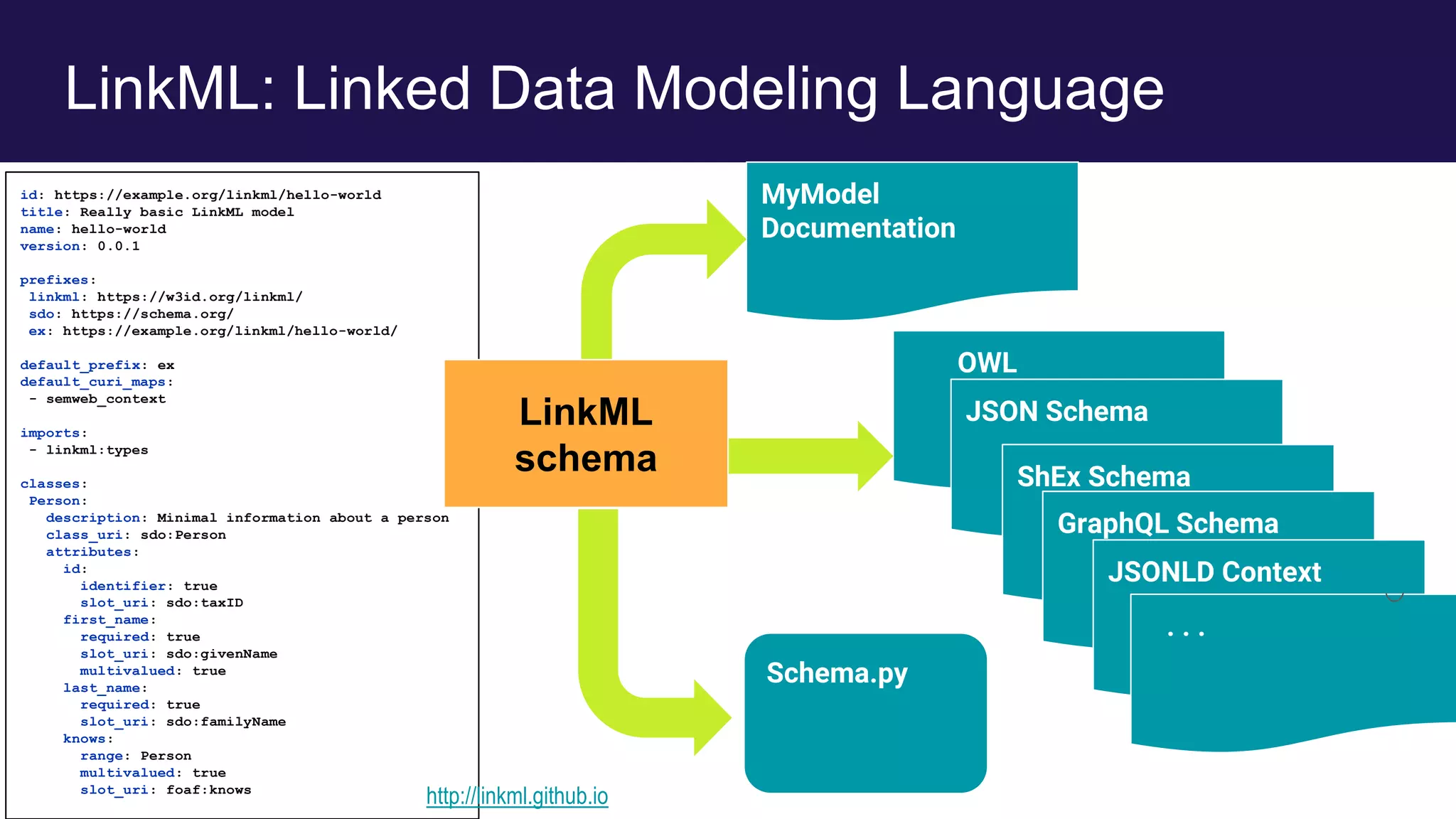
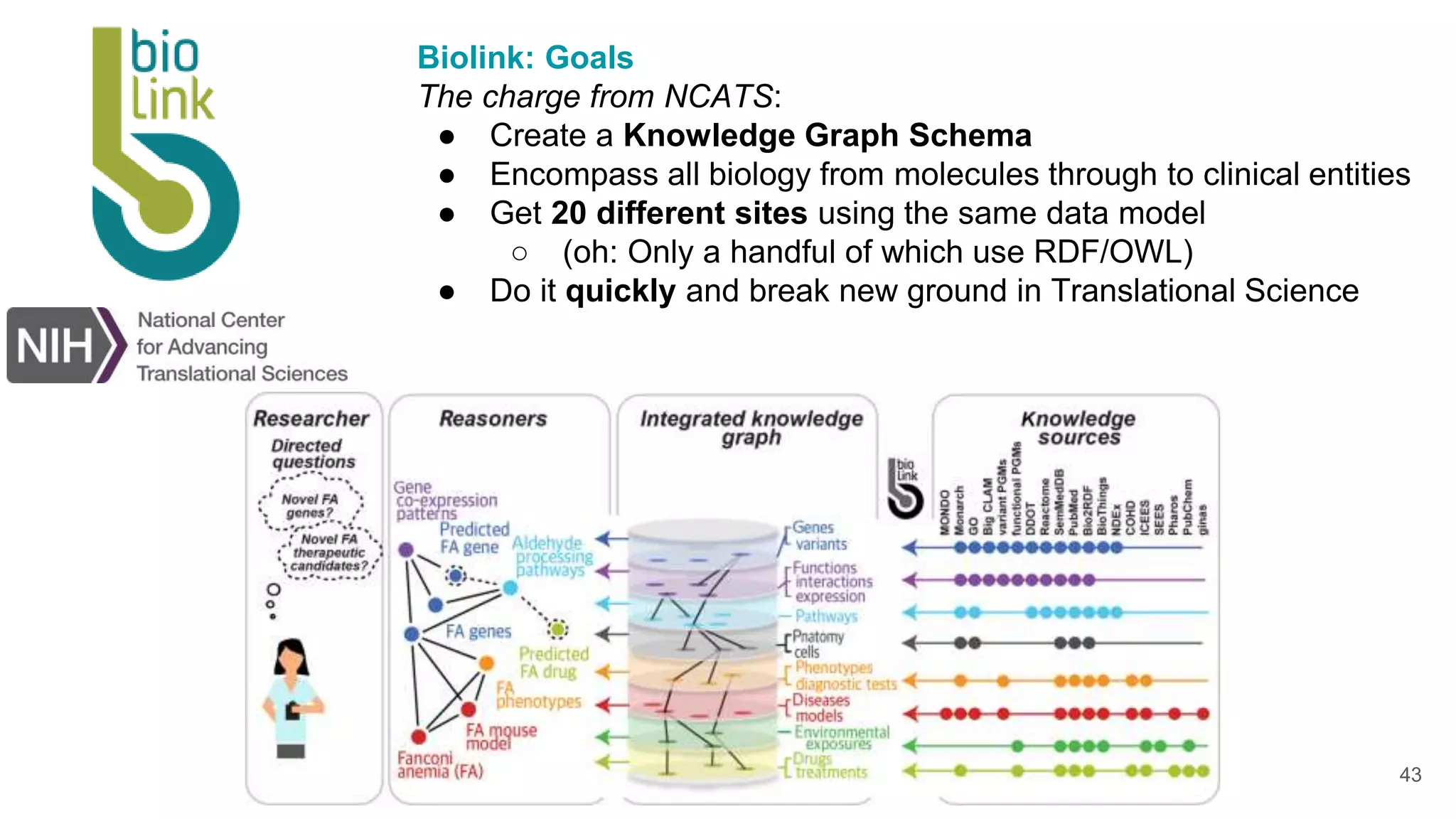
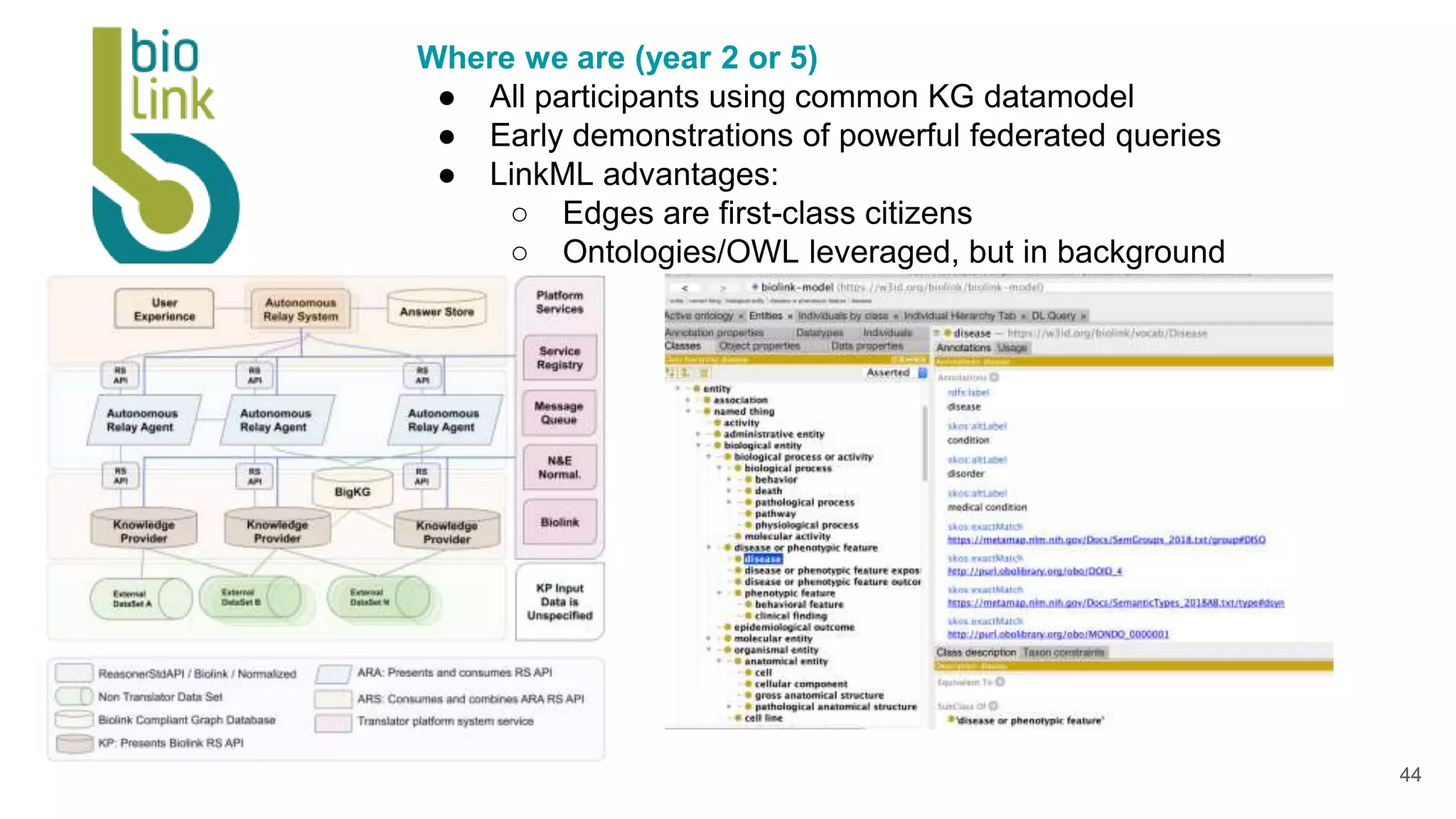
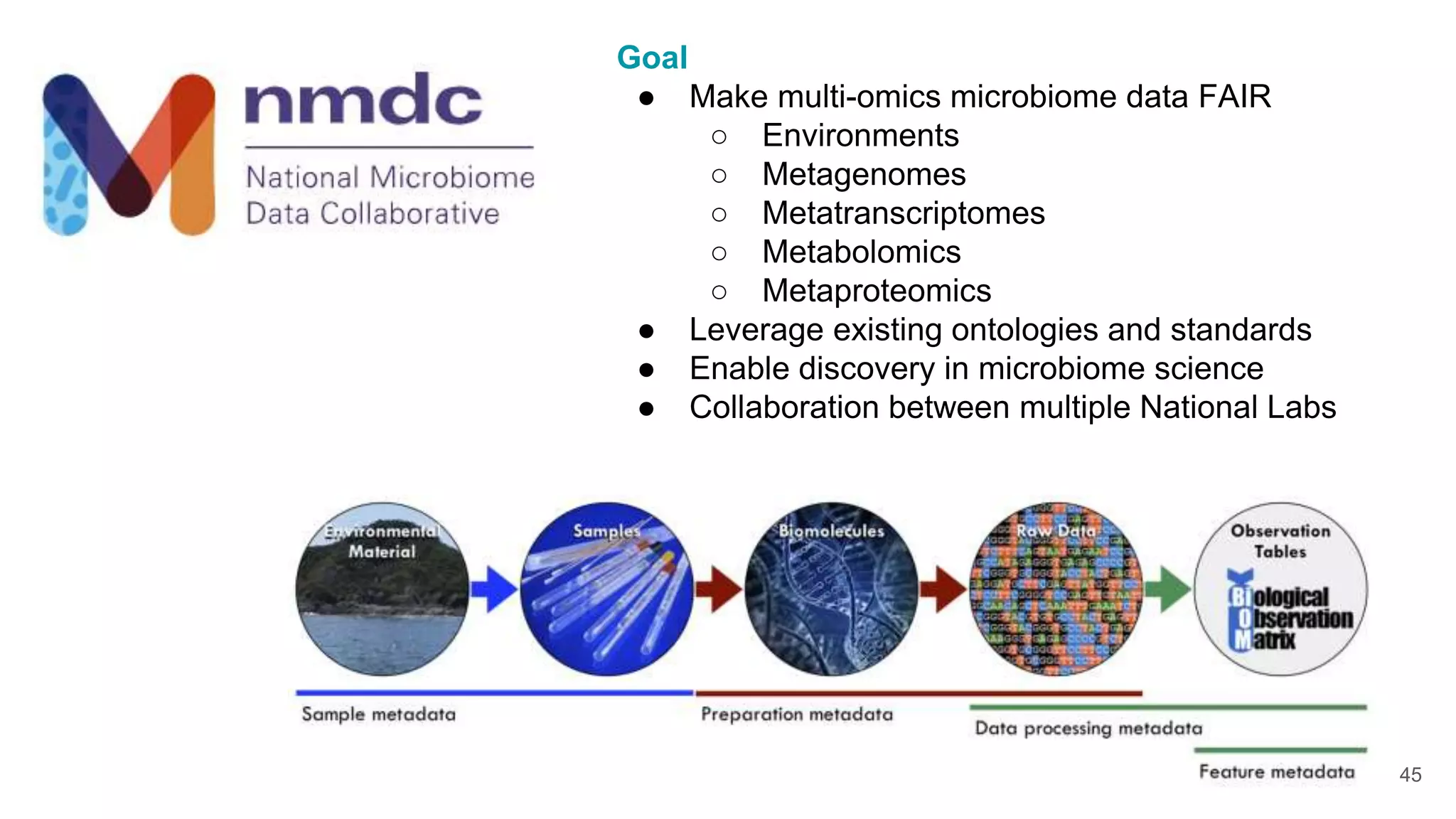
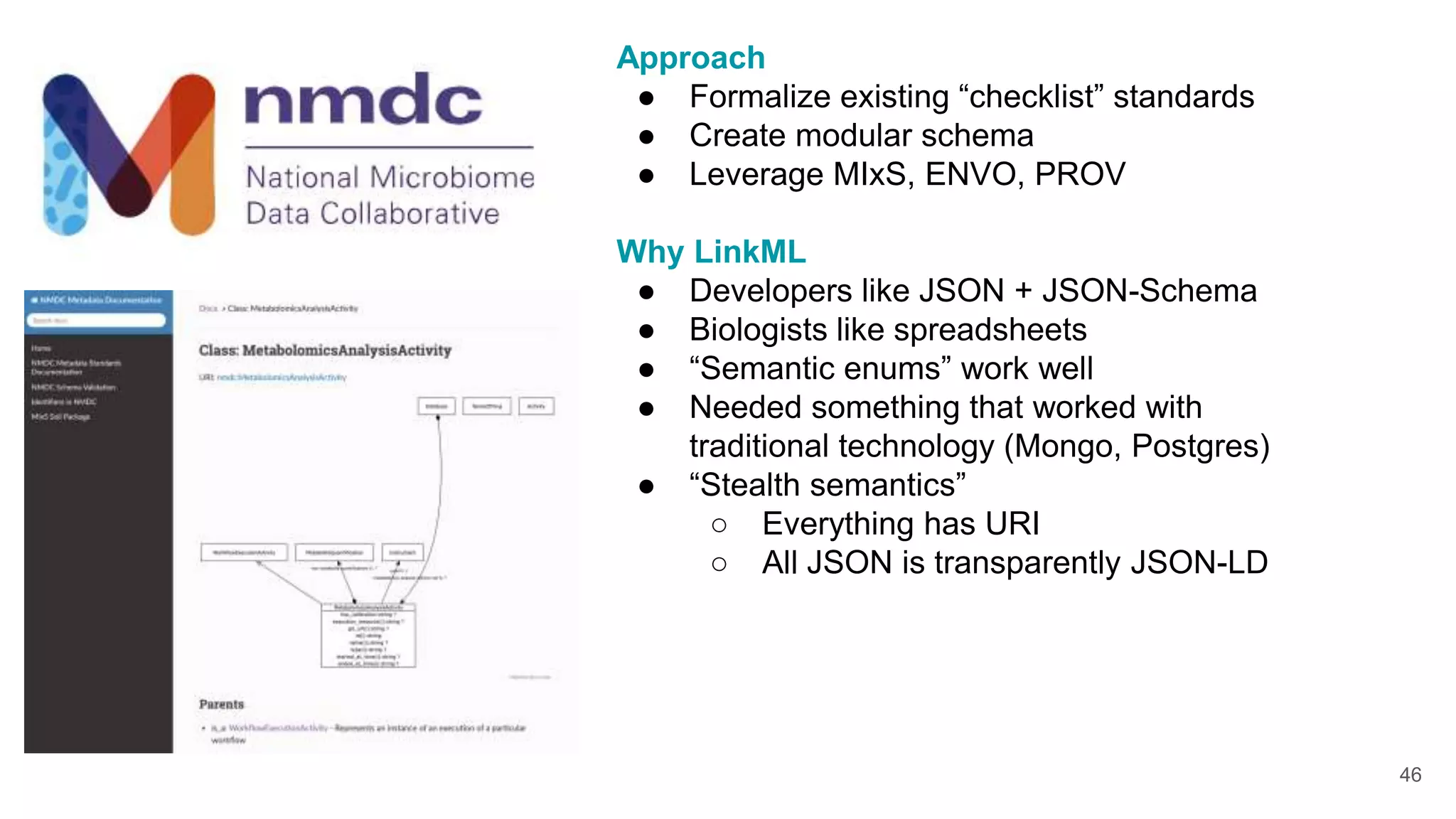
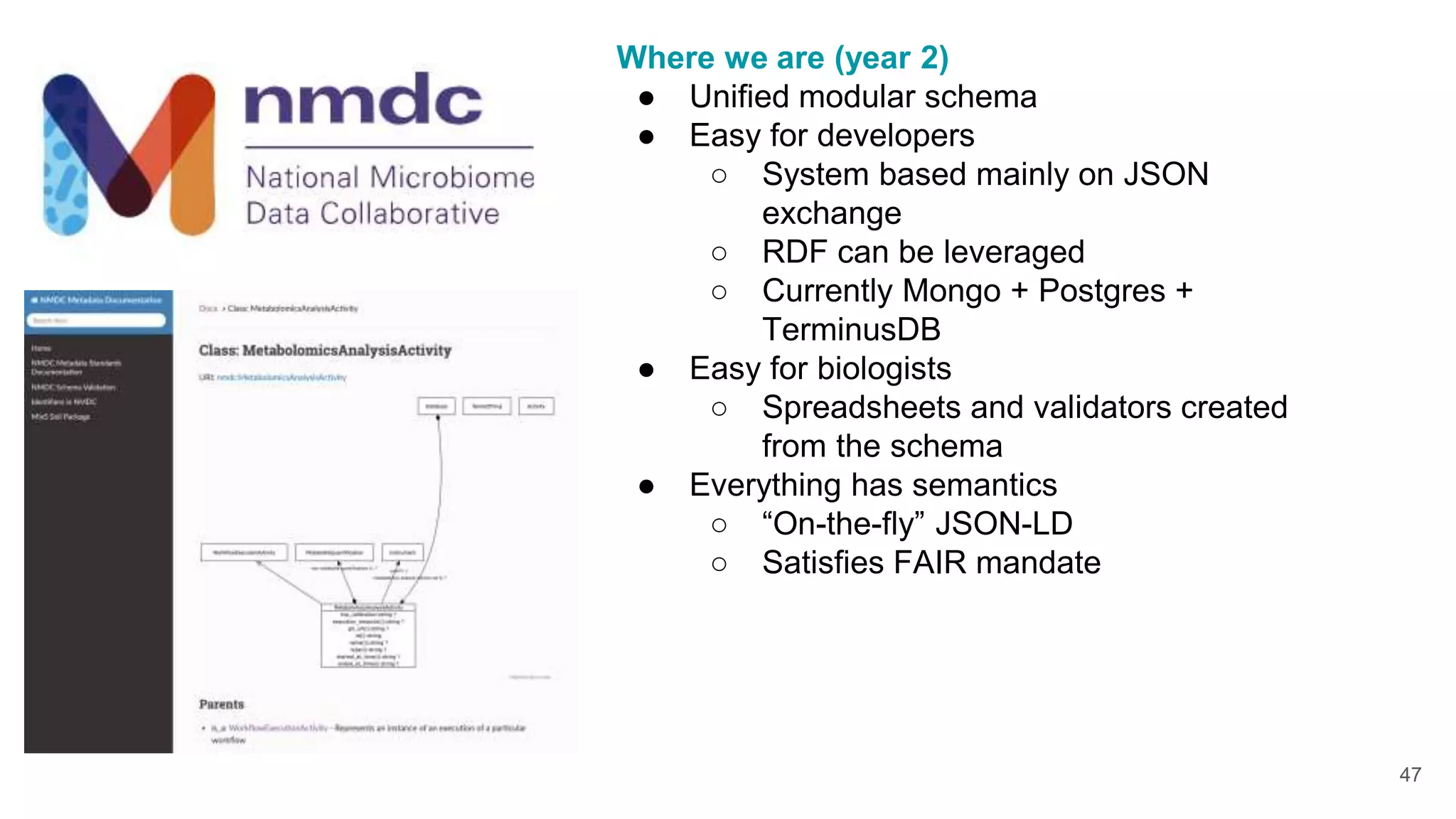
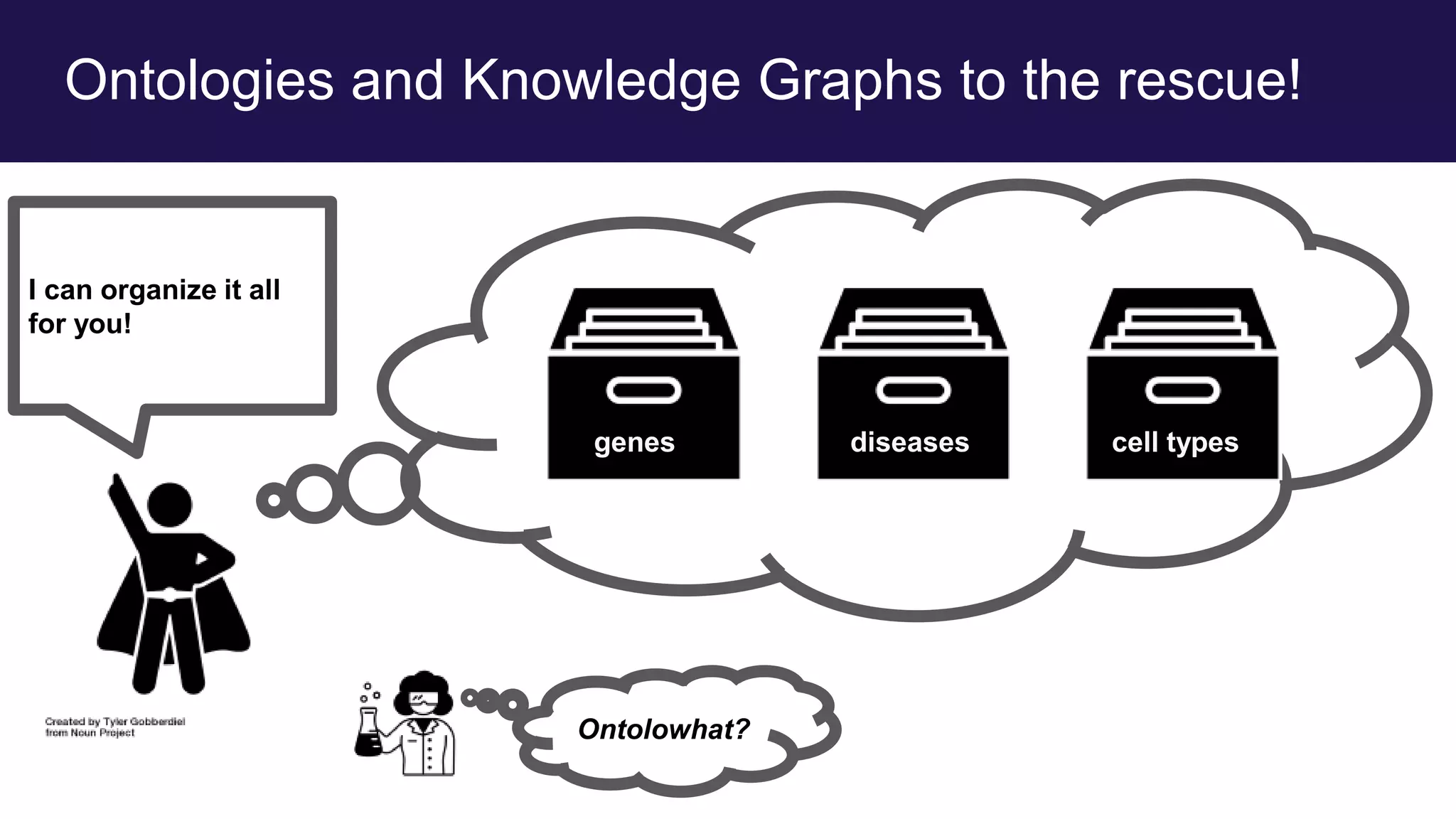
The document discusses collaboratively building a knowledge graph of life by connecting existing biological ontologies. It describes how ontologies can standardize and organize biological data by representing entities and their relationships in a graph. The challenges of integrating different ontology projects are addressed through initiatives like the Open Biological and Biomedical Ontologies (OBO) Foundry. The document outlines how ontologies can be formalized using OWL and connected using tools like the Ontology Development Kit to enable discovery across domains. Current efforts like the Gene Ontology, Biolink Model, and National Microbiome Data Collaborative are leveraging these techniques to create unified, semantically queryable knowledge graphs.





![Ontologies enable discovery
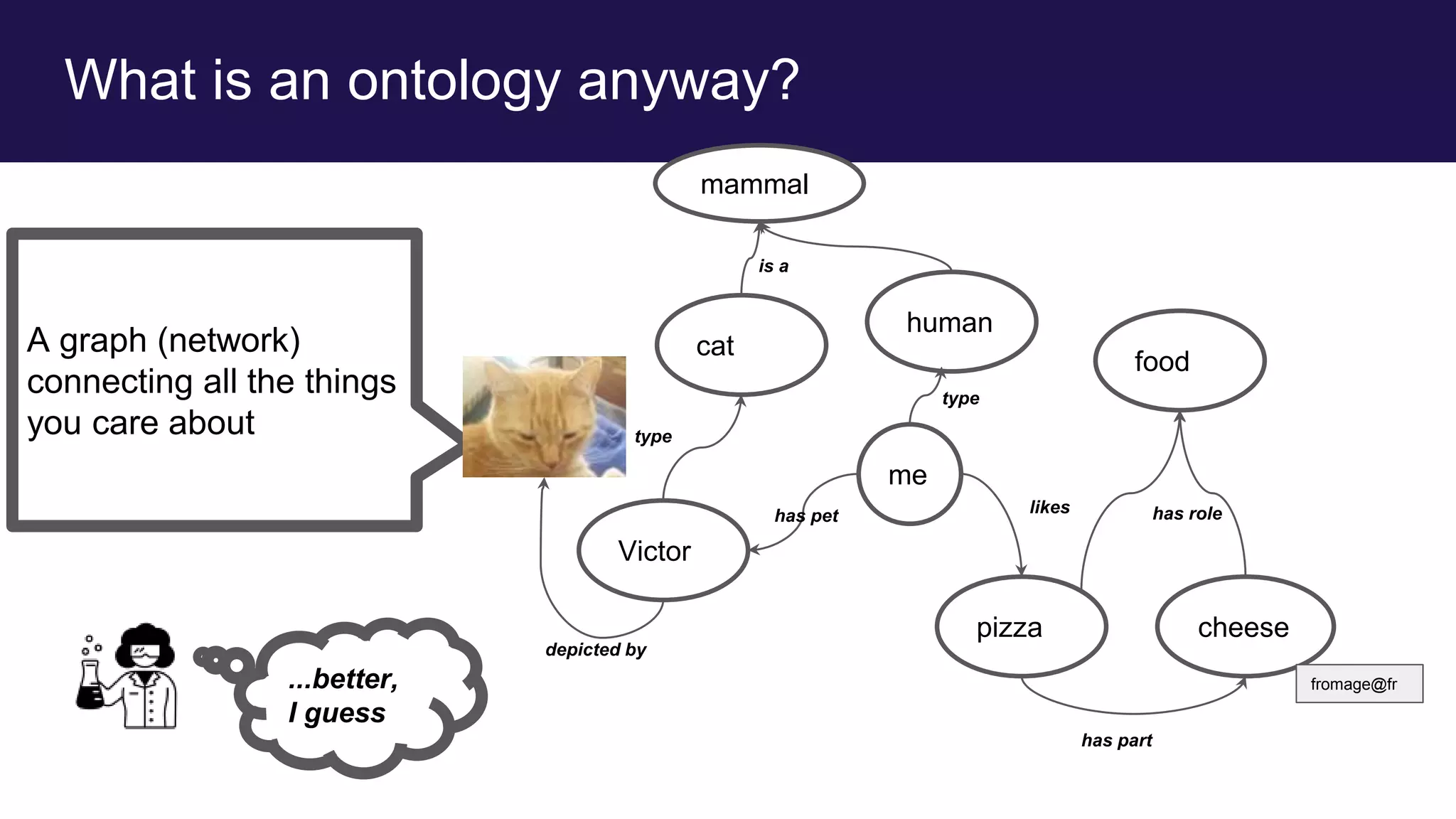
This is
fun
actually...
Do owners of different
kinds of pets like different
kinds of food? What do
those foods have in
common?
me
pizza cheese
food
Victor
cat
human
mammal
fromage@fr
type
is a
has pet
depicted by
likes
has part
has role
type
Lizard owners
like spicy food
[p=0.04]](https://image.slidesharecdn.com/obographgangtalk1-210702223118/75/Collaboratively-Creating-the-Knowledge-Graph-of-Life-12-2048.jpg)