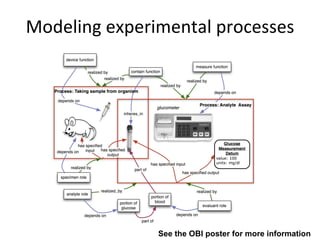
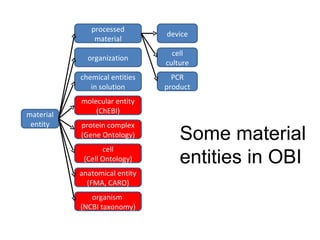
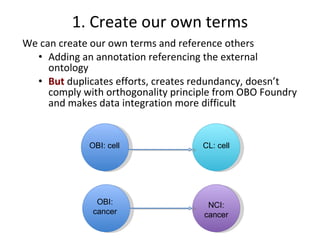
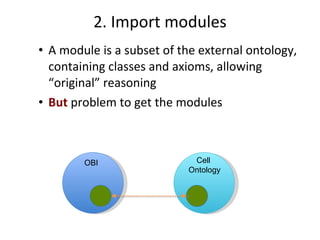
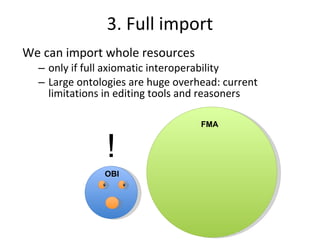
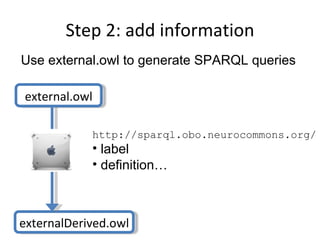
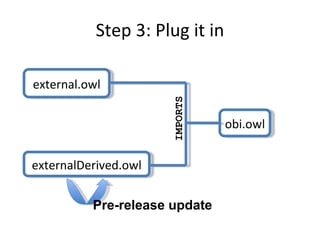
The document discusses MIREOT (Minimal information to reference external ontology terms), an approach used by the Ontology for Biomedical Investigations (OBI) project to import terms from external ontologies. It describes three approaches to importing terms - creating duplicate terms, importing modules, and full imports. It proposes importing only the classes needed using a minimal set of information to unambiguously identify terms from external ontologies. This process has been implemented in OBI and an online tool called OntoFox has been developed to facilitate the MIREOT process.