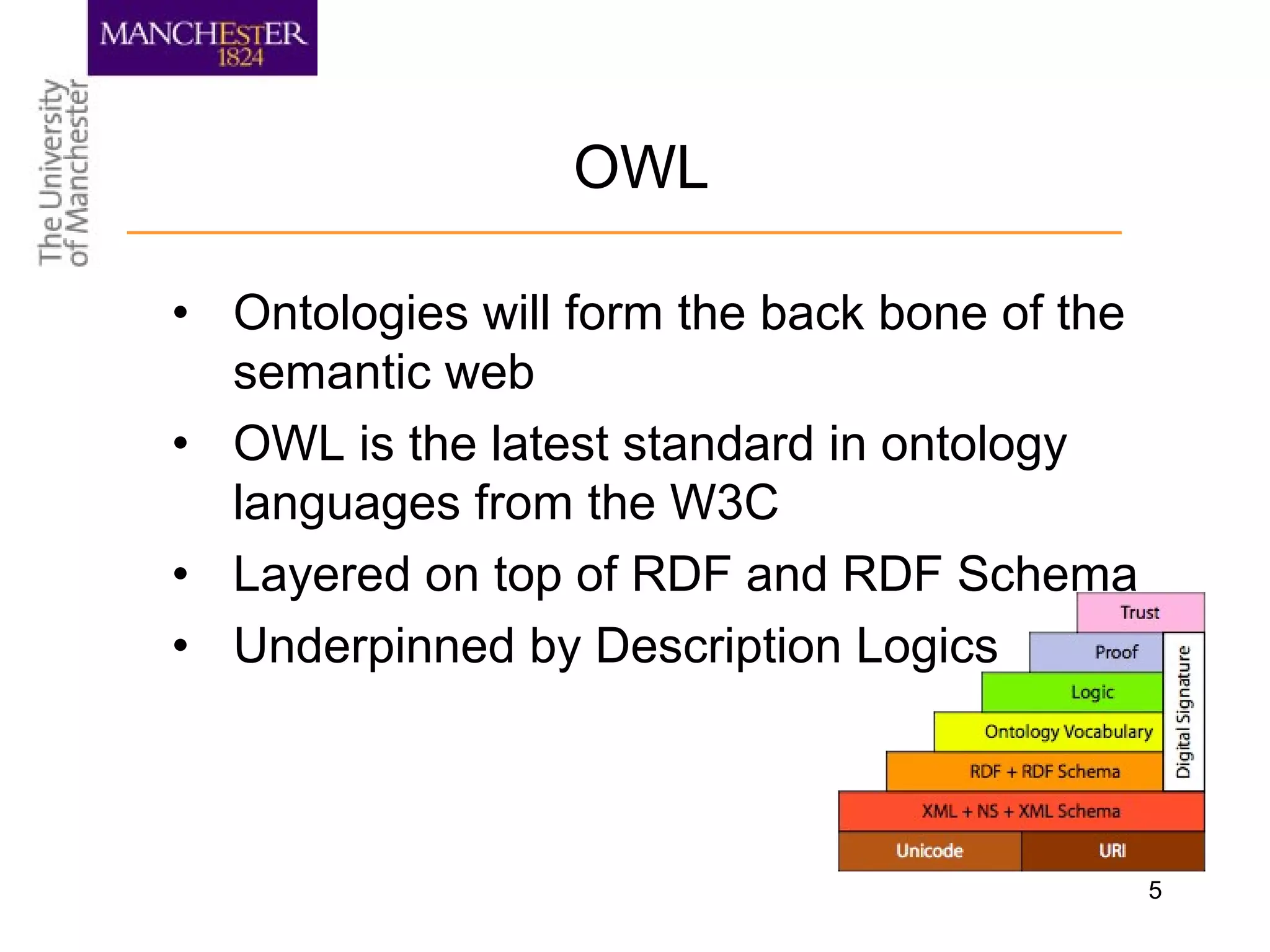
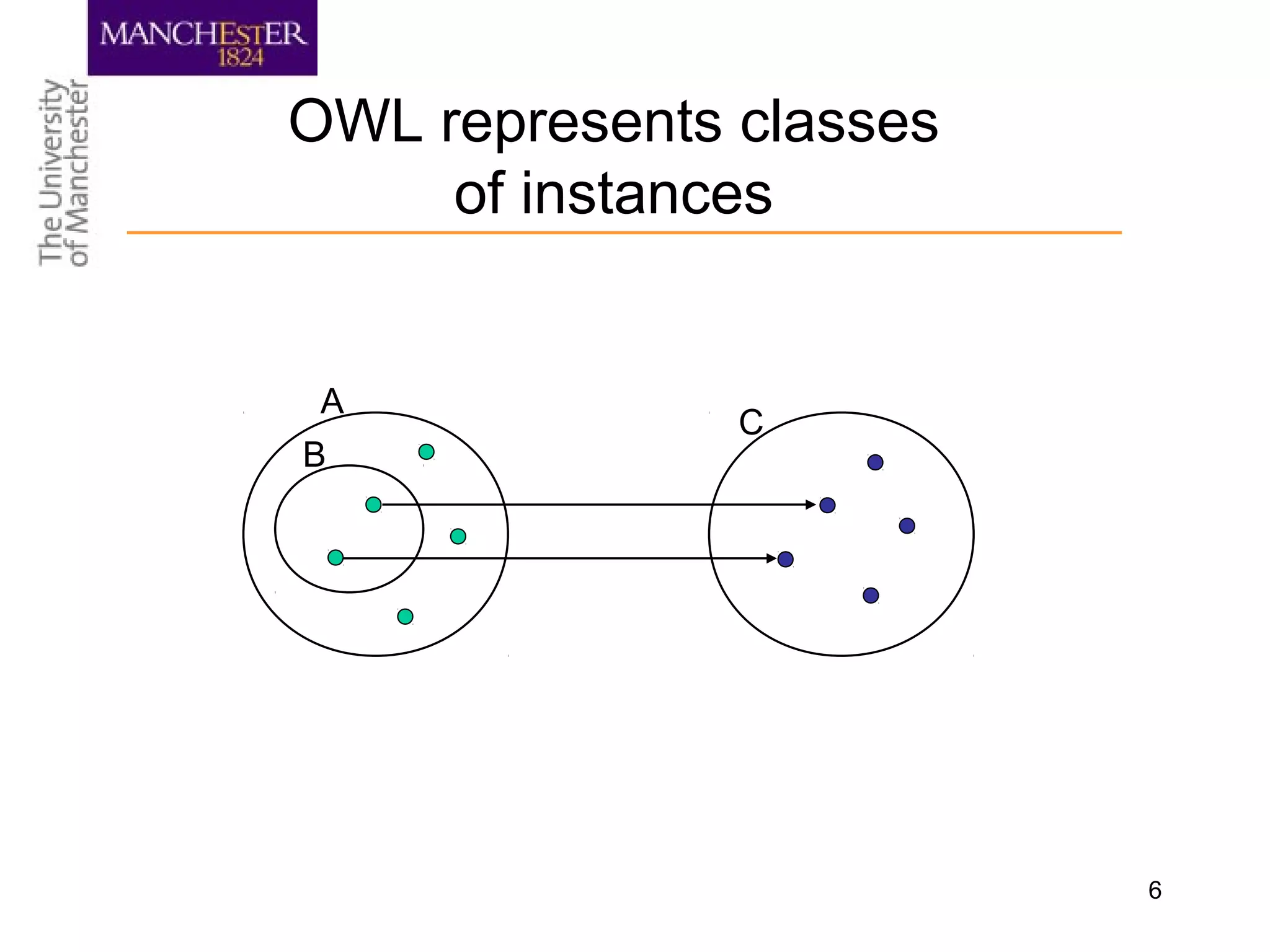
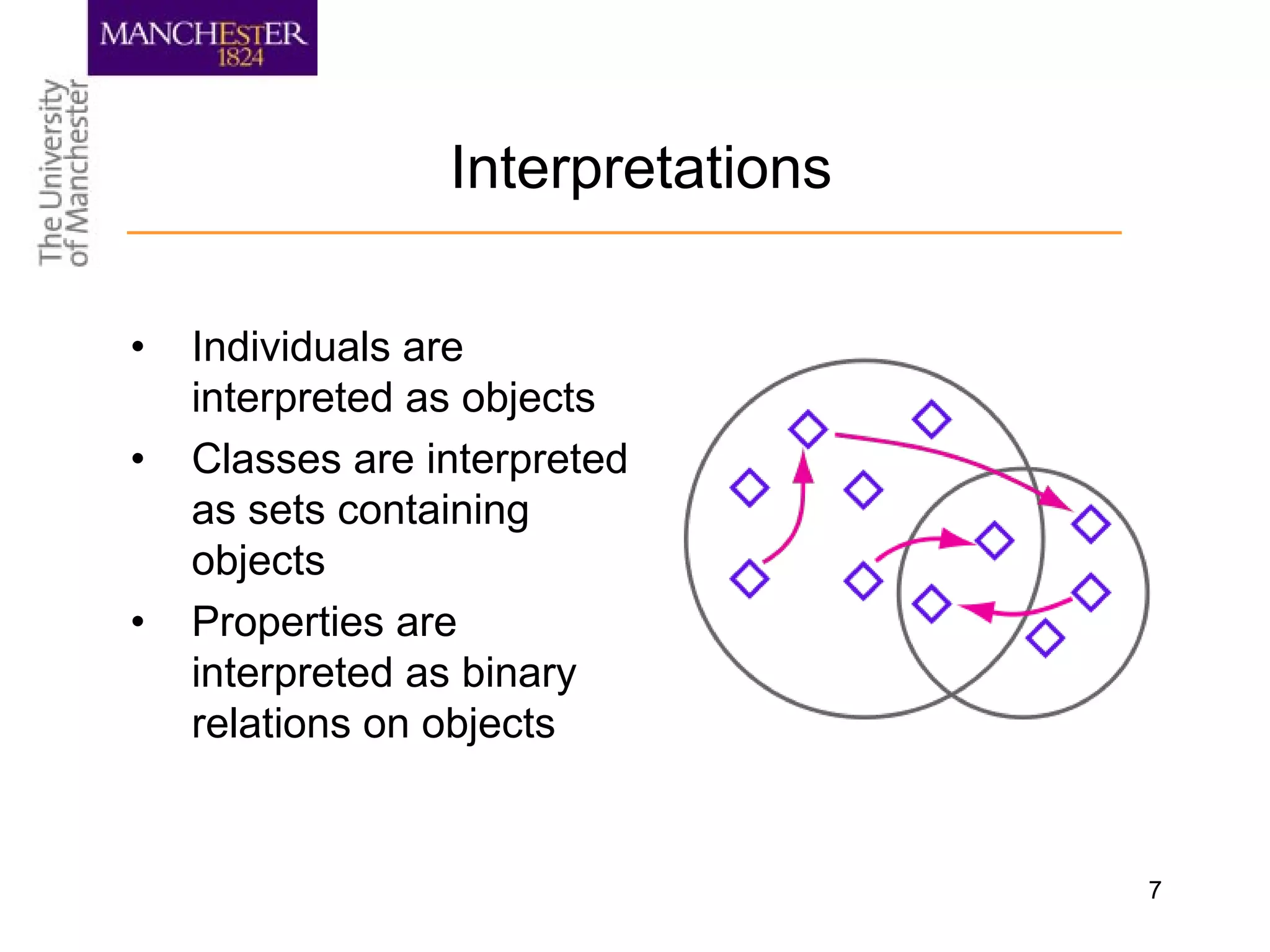
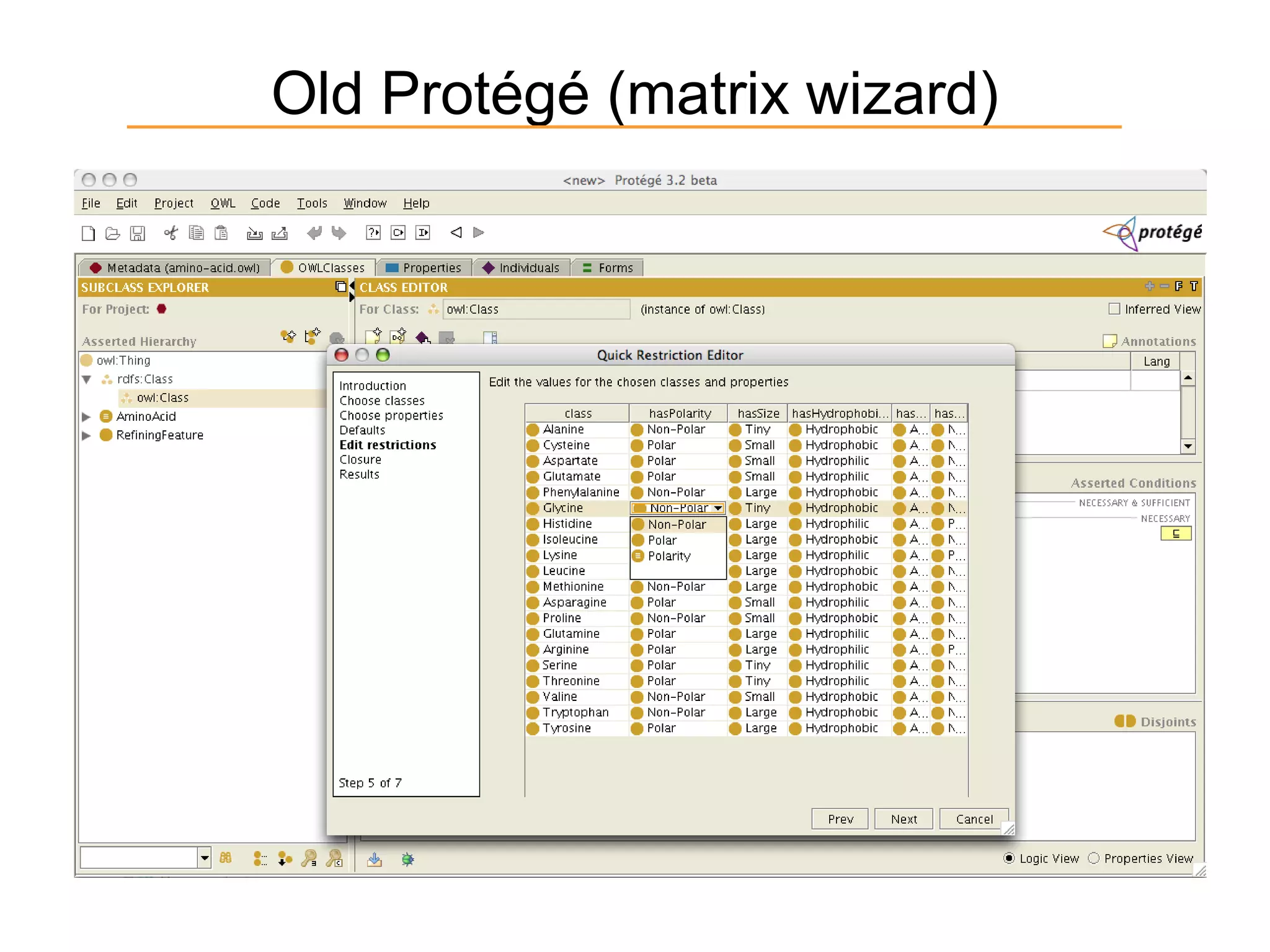
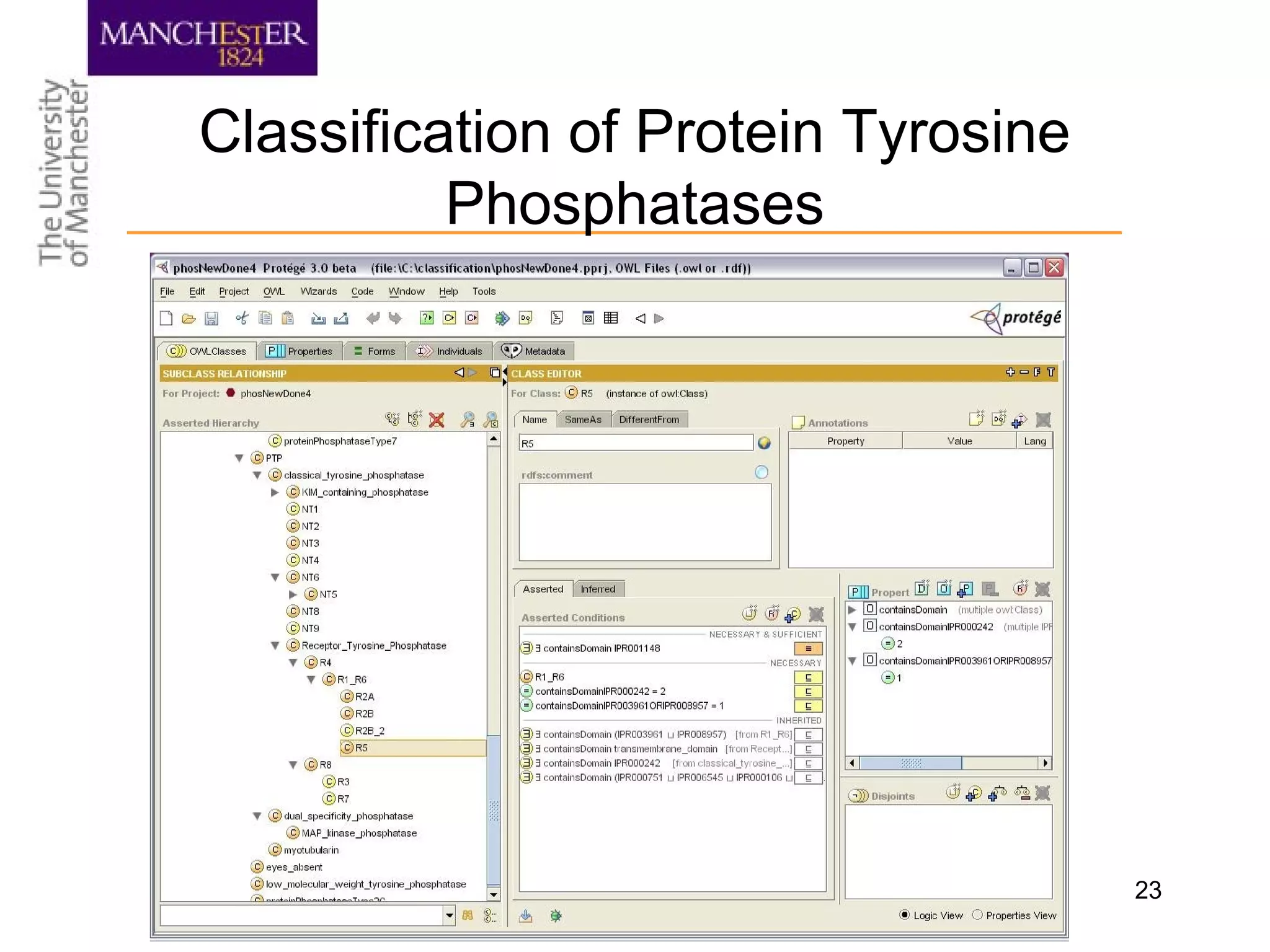
The document summarizes ontology research at the University of Manchester. It discusses defining ontologies using OWL, applying reasoning to classify proteins and detect missing knowledge, building ontologies collaboratively, and using ontologies to model biology and medicine. The research uses ontologies for tasks like protein classification, semantic search, and guiding data annotation in domains like genomics and healthcare.

![4
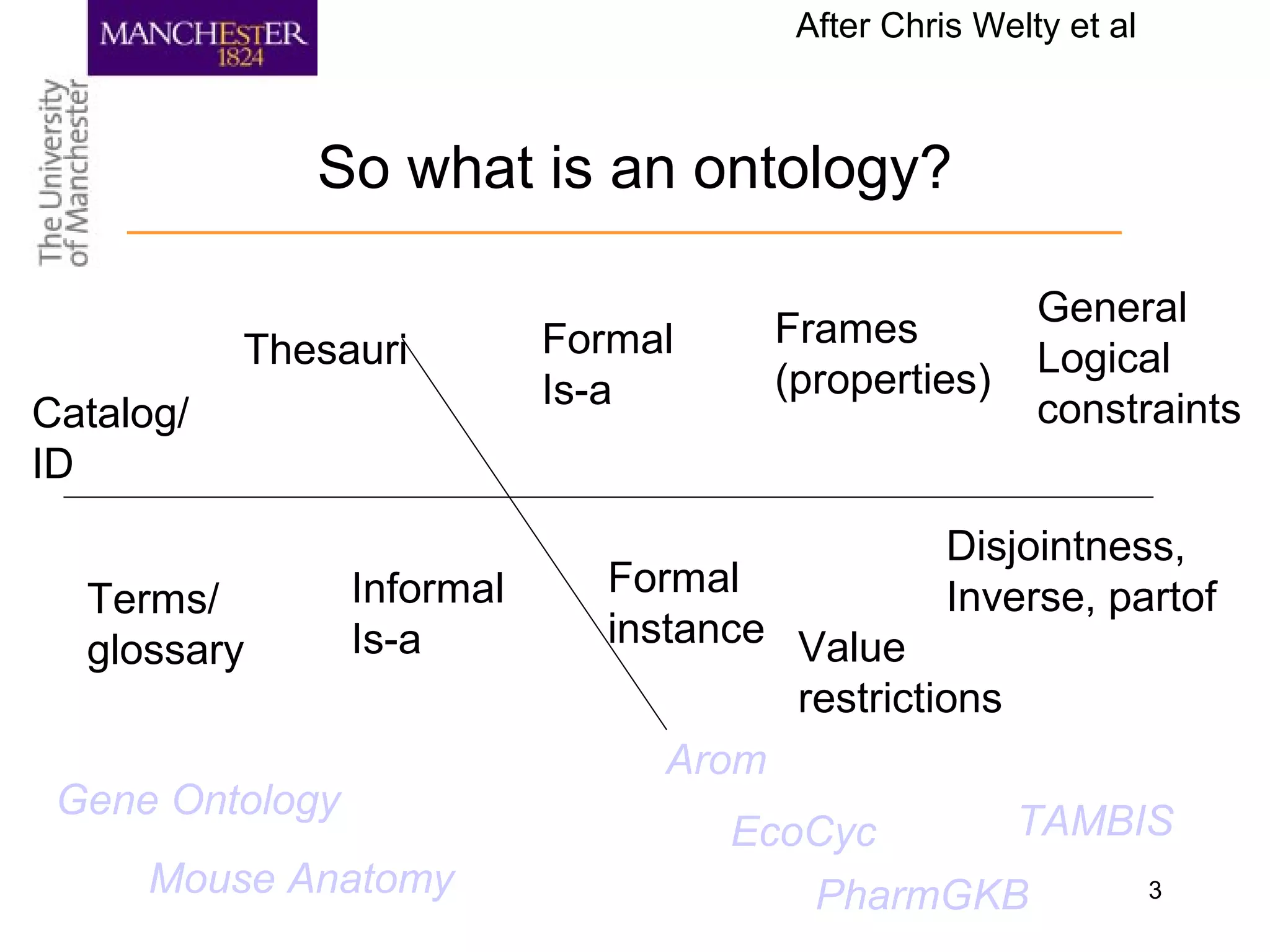
A Definition
o a set of logical axioms designed to account for the
intended meaning of a formal vocabulary used to describe
a certain (conceptualisation of) reality [Guarino 1998]
o “conceptualisation of” inserted by me
o “Logical axioms” means a formal definition of meaning of
terms in a formal language
o Formal language—something a computer an reason with
o Use symbols to make inferences
o Symbols represent things and their relationships
o Making inferences about things computationally amenable](https://image.slidesharecdn.com/s7zpzpptjci0evb5h88p-signature-f84076d802018ccc31f808b4d674dd5edfbd24357d894cb2420d389f15b69e02-poli-140716122622-phpapp02/75/Ontology-at-Manchester-4-2048.jpg)