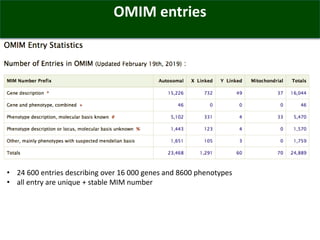
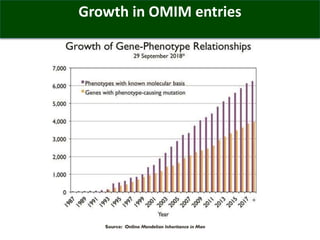
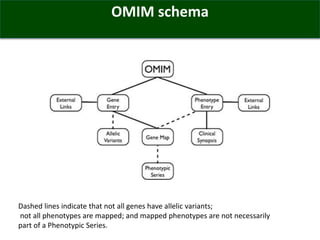
The OMIM database provides structured summaries of the relationship between human genotypes and phenotypes by reviewing the biomedical literature. It was initiated in the 1960s as MIM and became an online database called OMIM in 1985. OMIM contains over 24,600 entries describing more than 16,000 genes and 8,600 phenotypes. The entries are updated nightly and provide a structured format to describe genotype-phenotype relationships along with interactive tools like genome coordinate searching and phenotypic series.