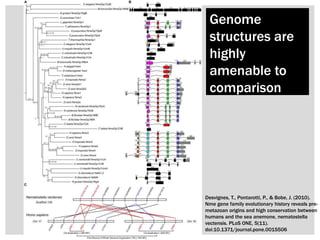
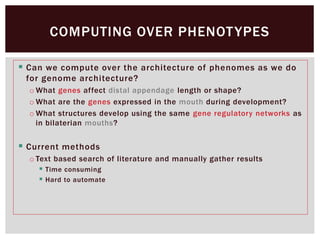
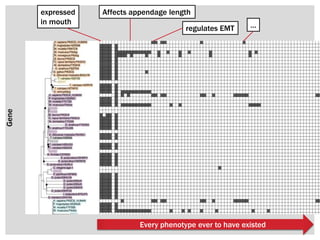
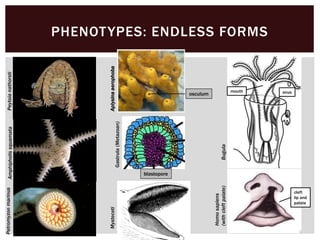
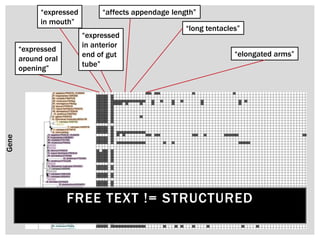
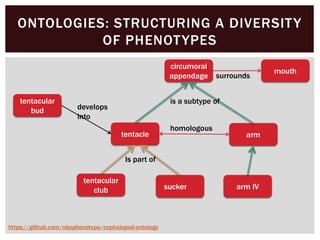
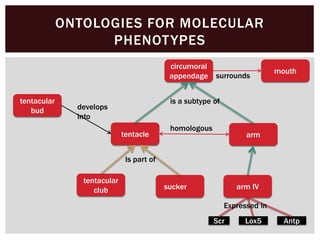
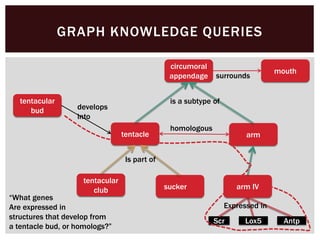
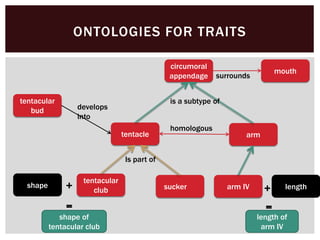
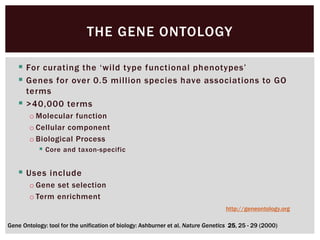
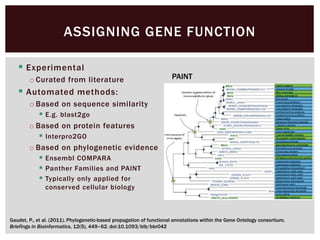
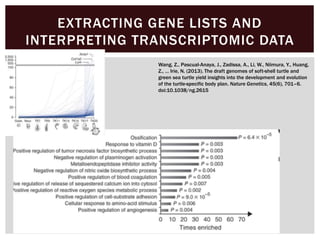
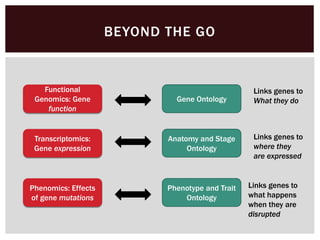
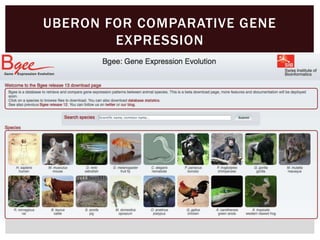
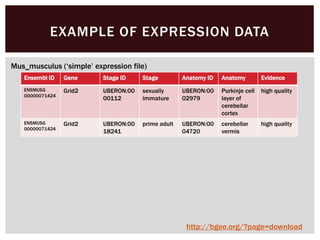
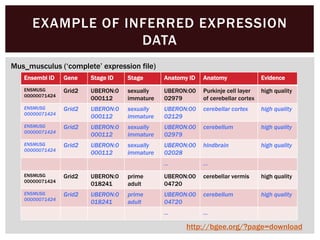
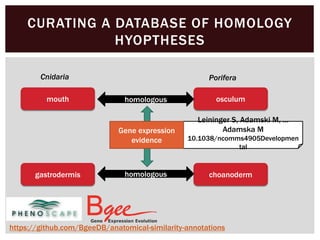
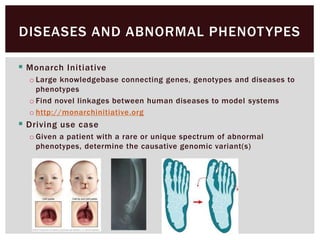
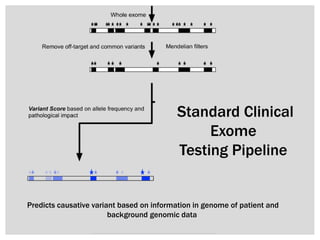
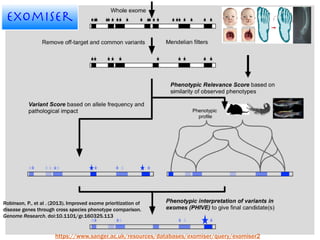
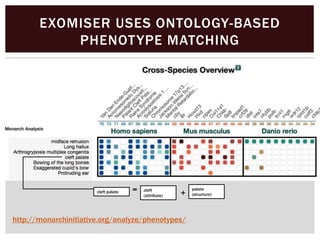
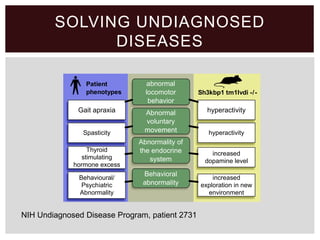
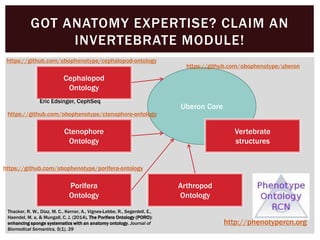
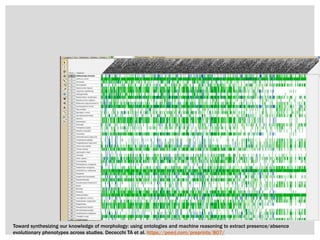
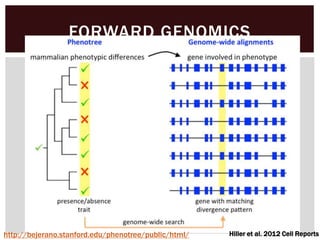
The document discusses the integration of genetic and phenotypic data through the use of ontologies, emphasizing the importance of structuring phenotype information for comparative biology. It highlights various ontological tools and databases that facilitate the analysis of gene expression, evolutionary relationships, and the application of standardized vocabularies for research. Key resources mentioned include the Gene Ontology, Uberon, and Phenoscape, which are pivotal for organizing biological knowledge across species.