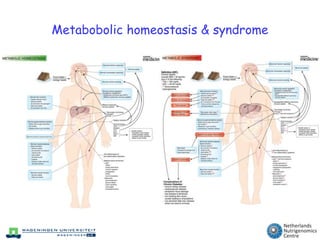
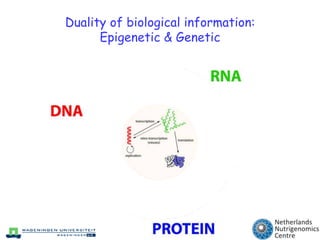
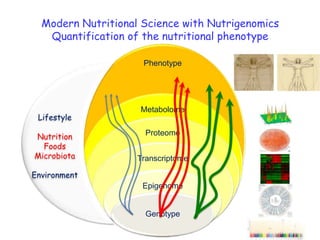
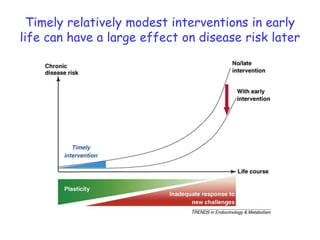
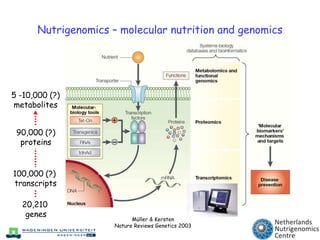
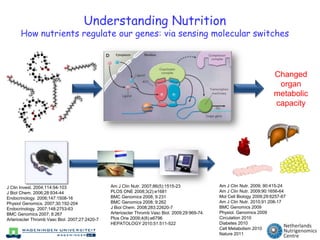
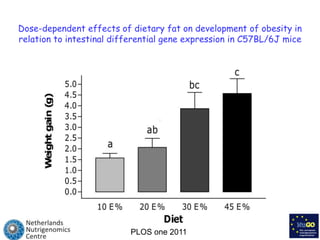
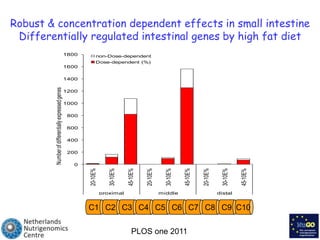
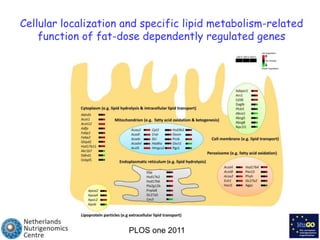
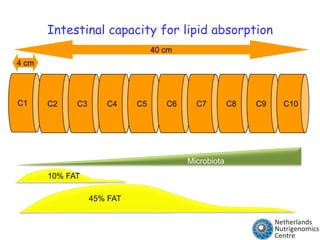
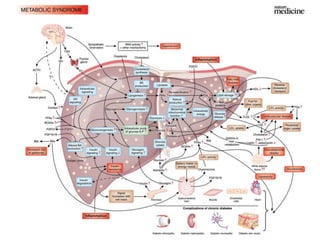
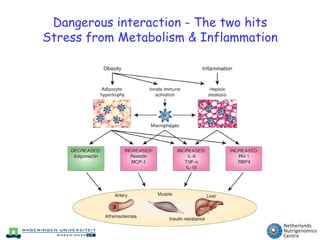
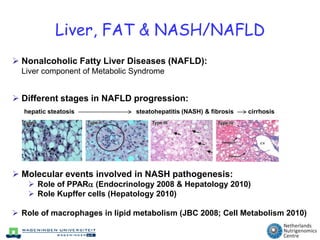
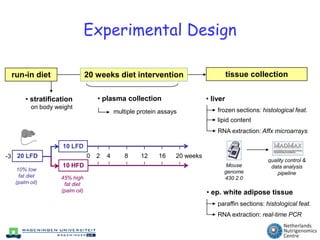
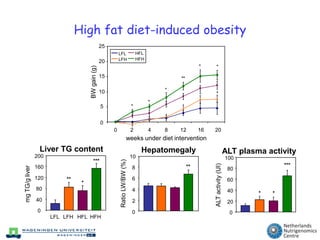
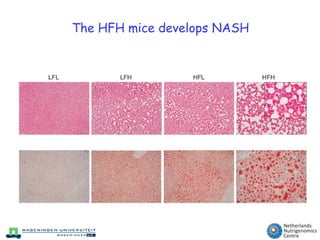
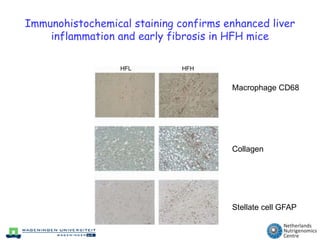
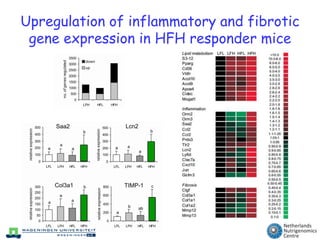
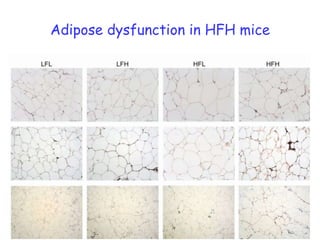
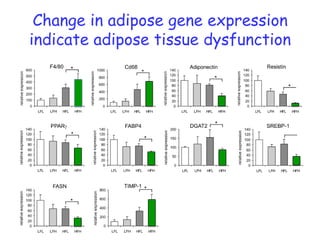
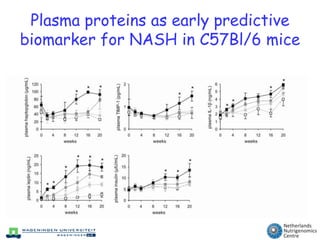
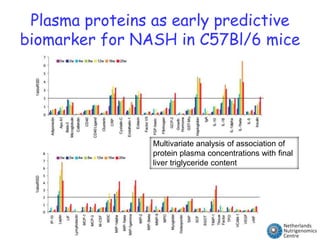
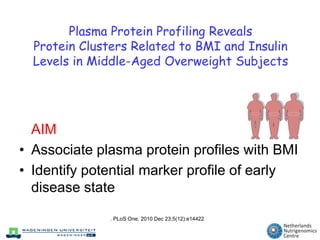
The document discusses the application of nutrigenomics in understanding metabolic homeostasis and the plasticity of health influenced by nutrition and lifestyle. It emphasizes the significance of personalized genomes, the impact of nutrition on gene regulation, and the roles of metabolites and proteins in metabolic diseases, particularly nonalcoholic fatty liver disease. The research highlights the interconnectedness of adipose tissue dysfunction and liver health, proposing potential biomarkers for disease progression and the importance of maintaining metabolic flexibility through lifestyle interventions.