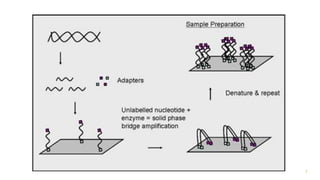
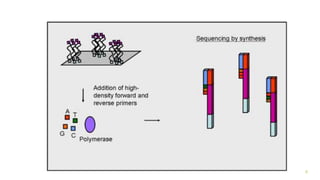
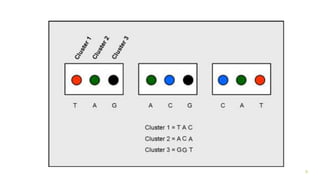
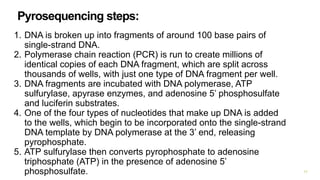
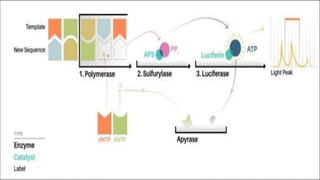
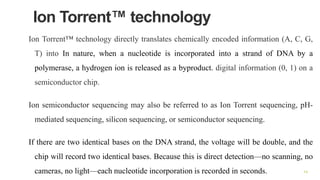
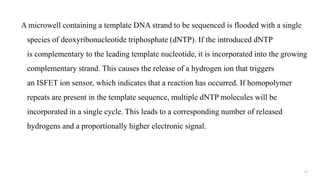
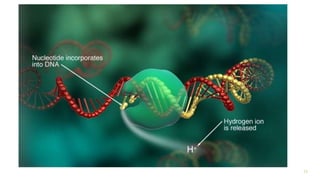
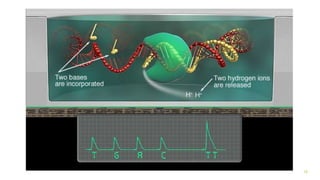
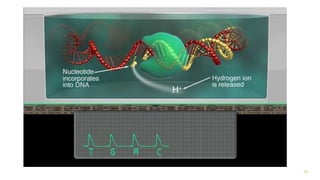
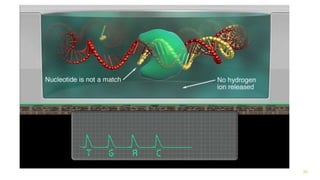
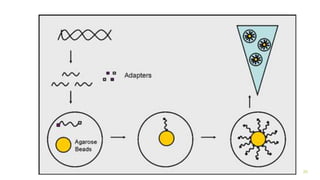
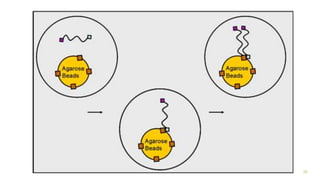
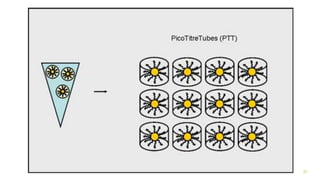
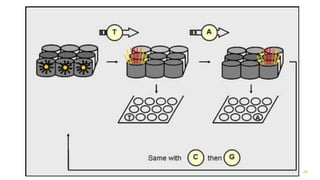
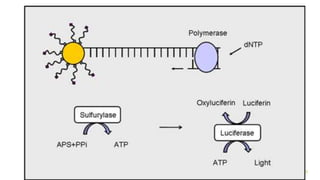
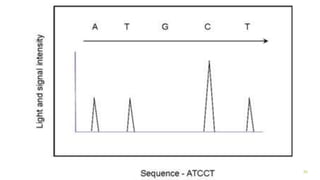
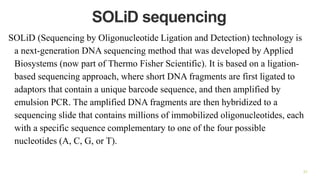
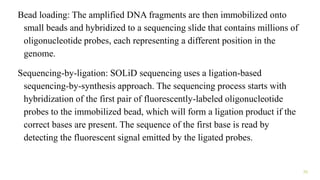
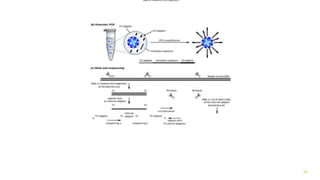
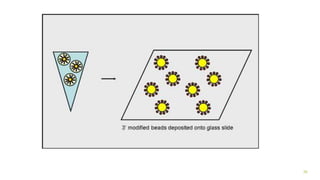
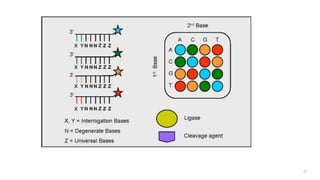
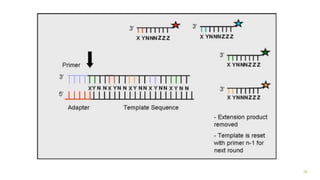
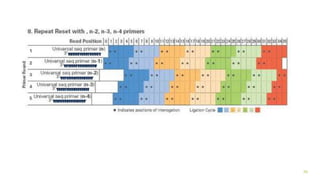
The document outlines various next-generation sequencing strategies, including Illumina, pyrosequencing, ion torrent technology, and 454 sequencing, detailing their processes from DNA library preparation to data analysis. It emphasizes the significance of base calling and data interpretation in extracting meaningful insights from the sequencing data. Applications for sequencing technology span across medicine, agriculture, and biotechnology, indicating its potential for future innovations such as precision medicine and gene therapy.