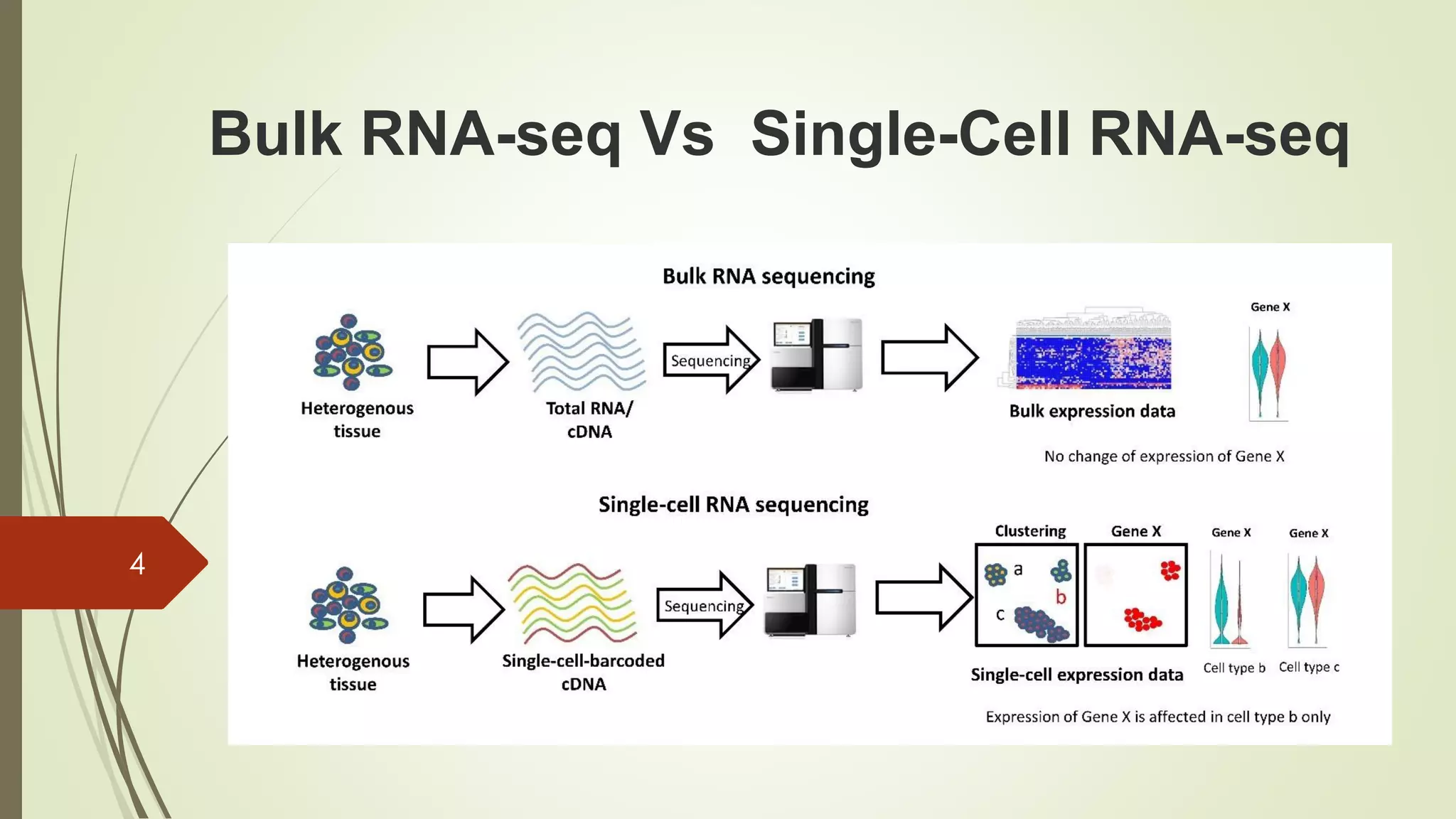
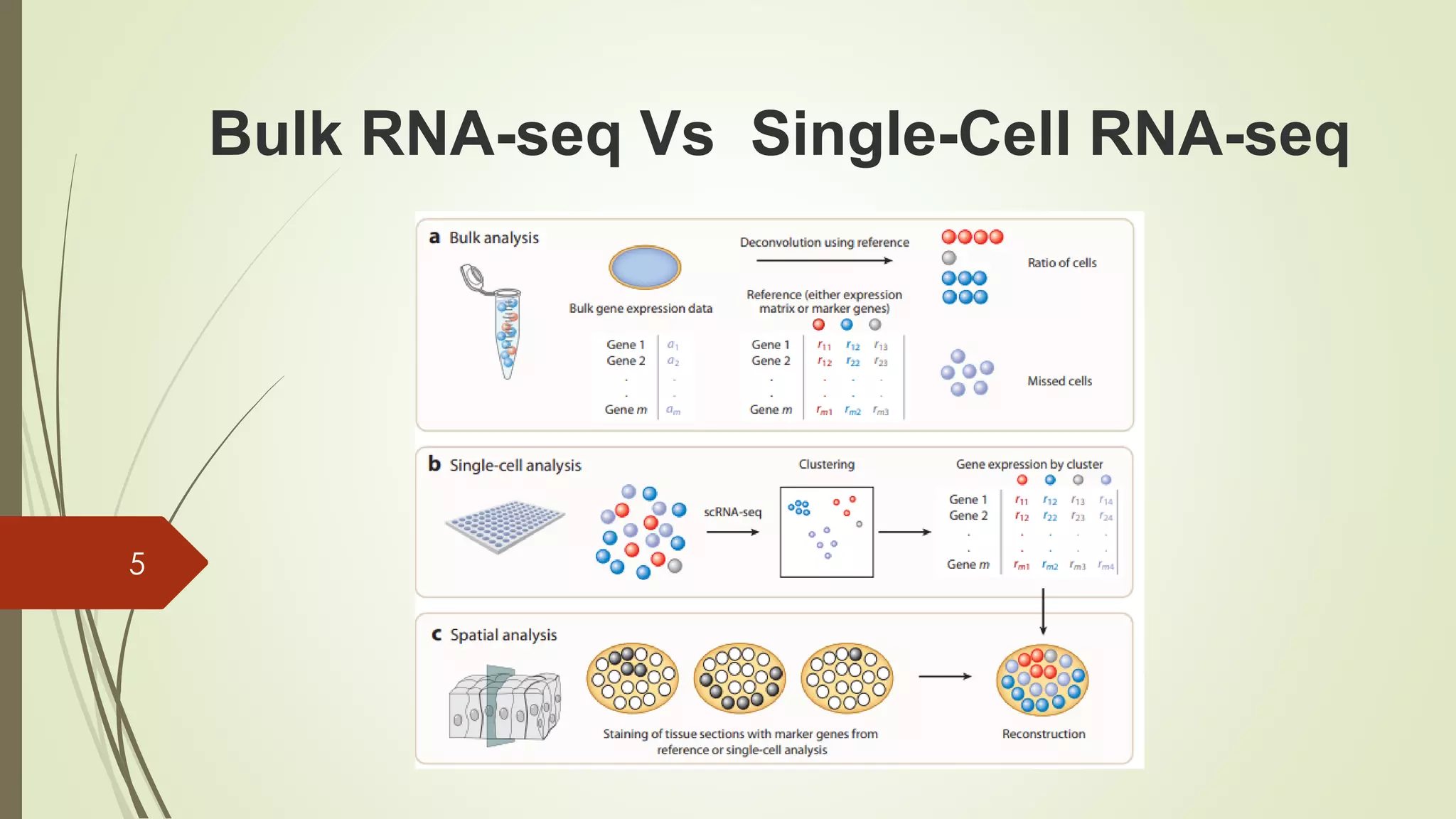
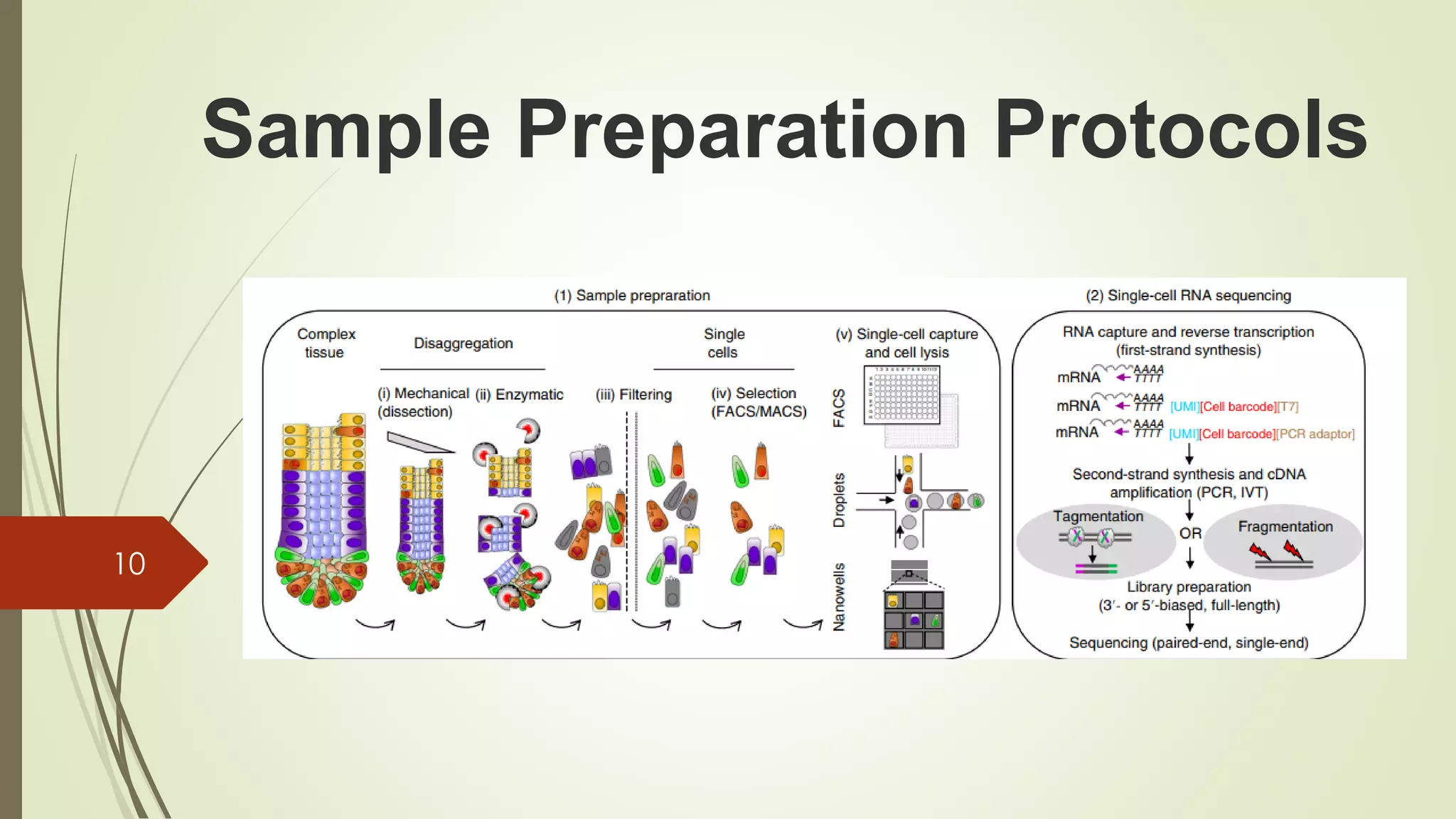
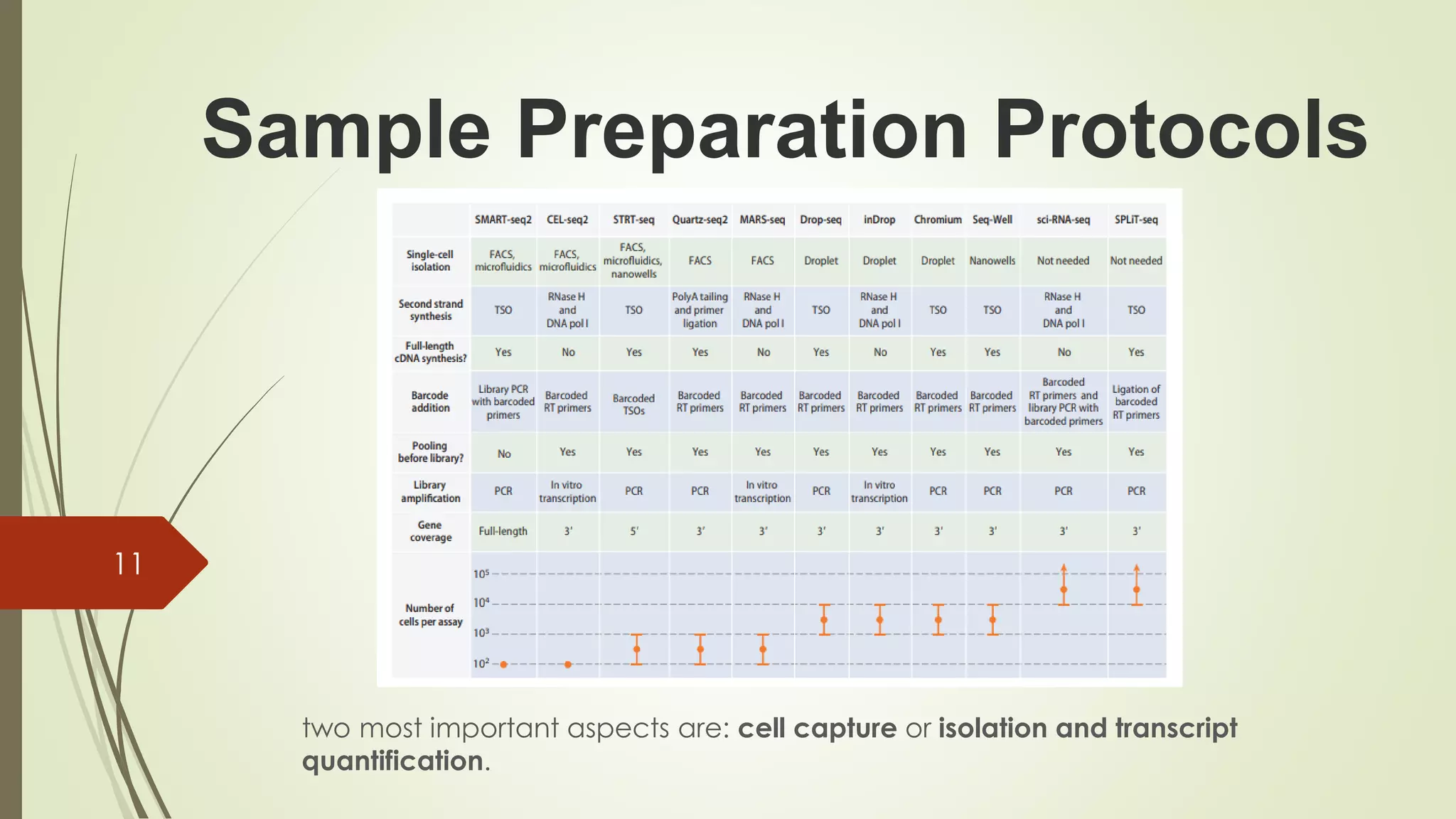
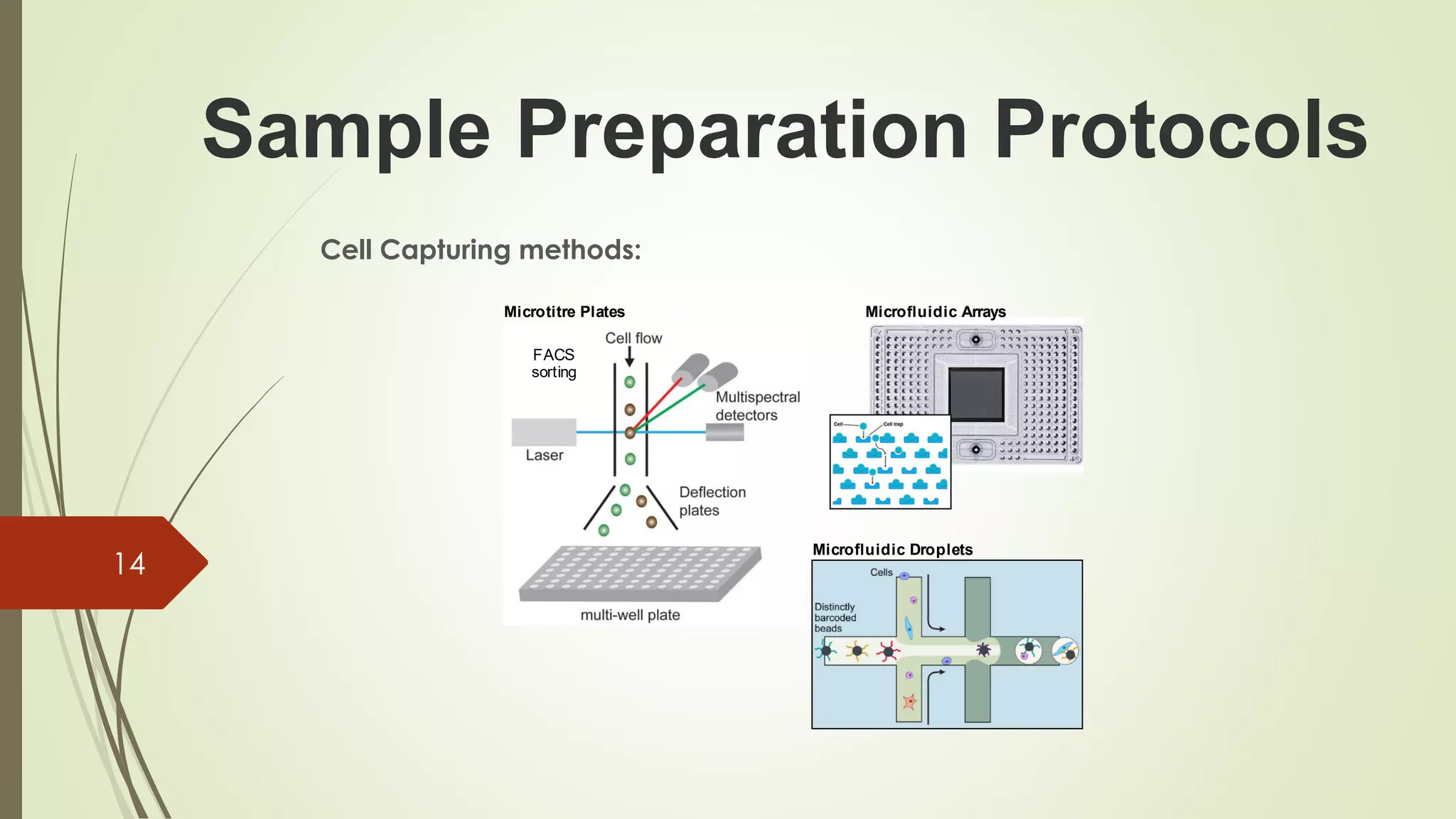
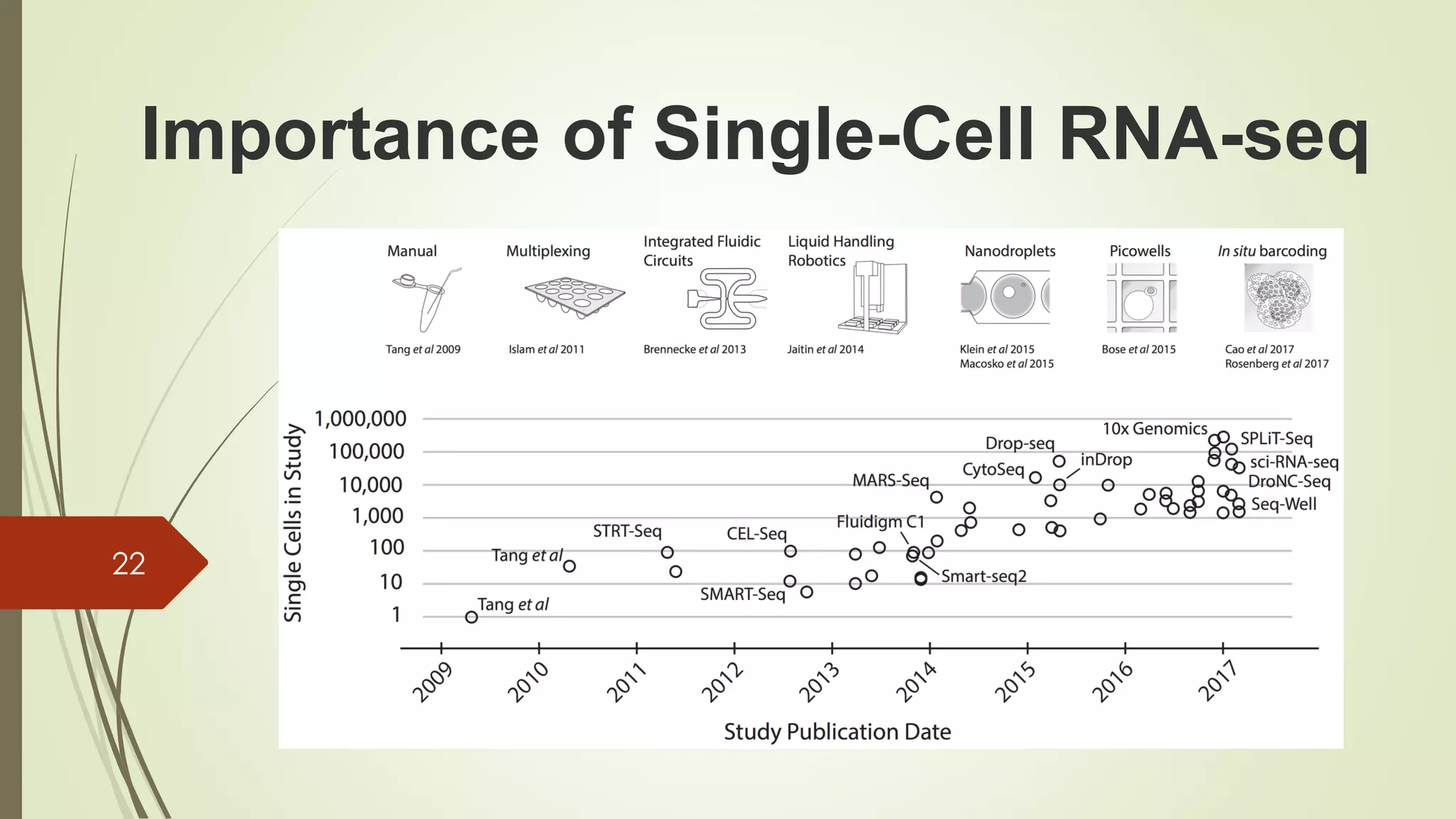
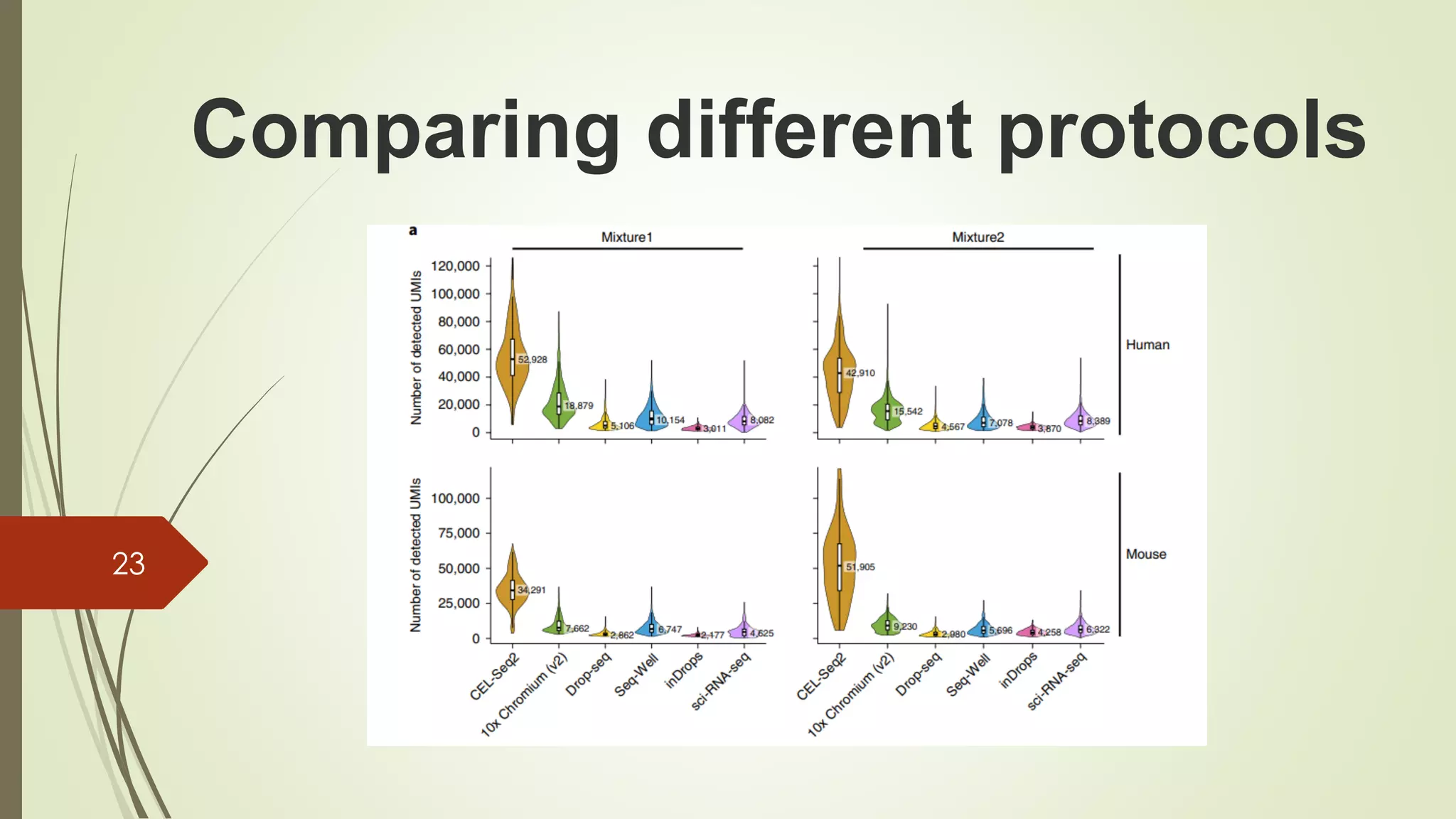
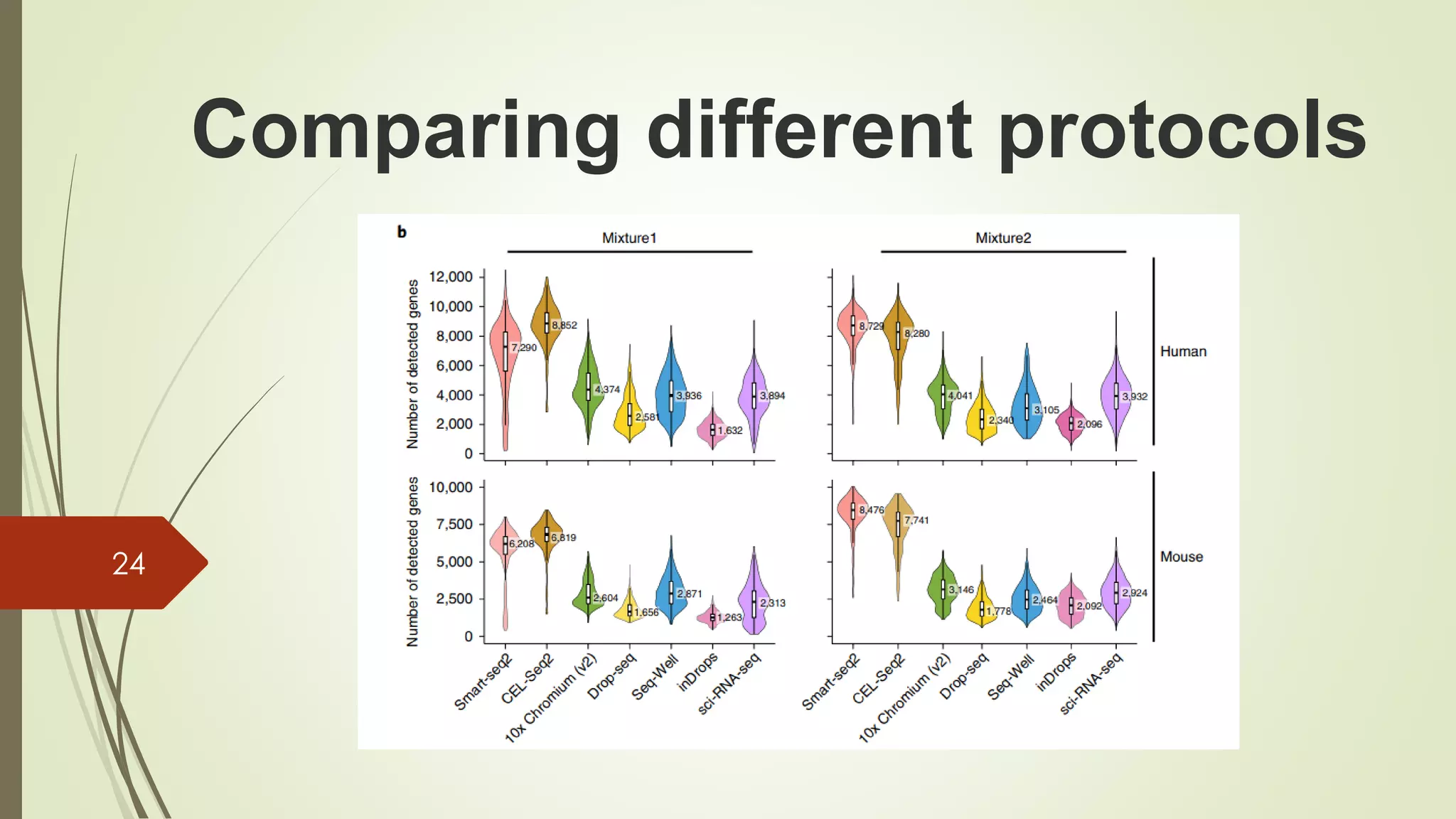
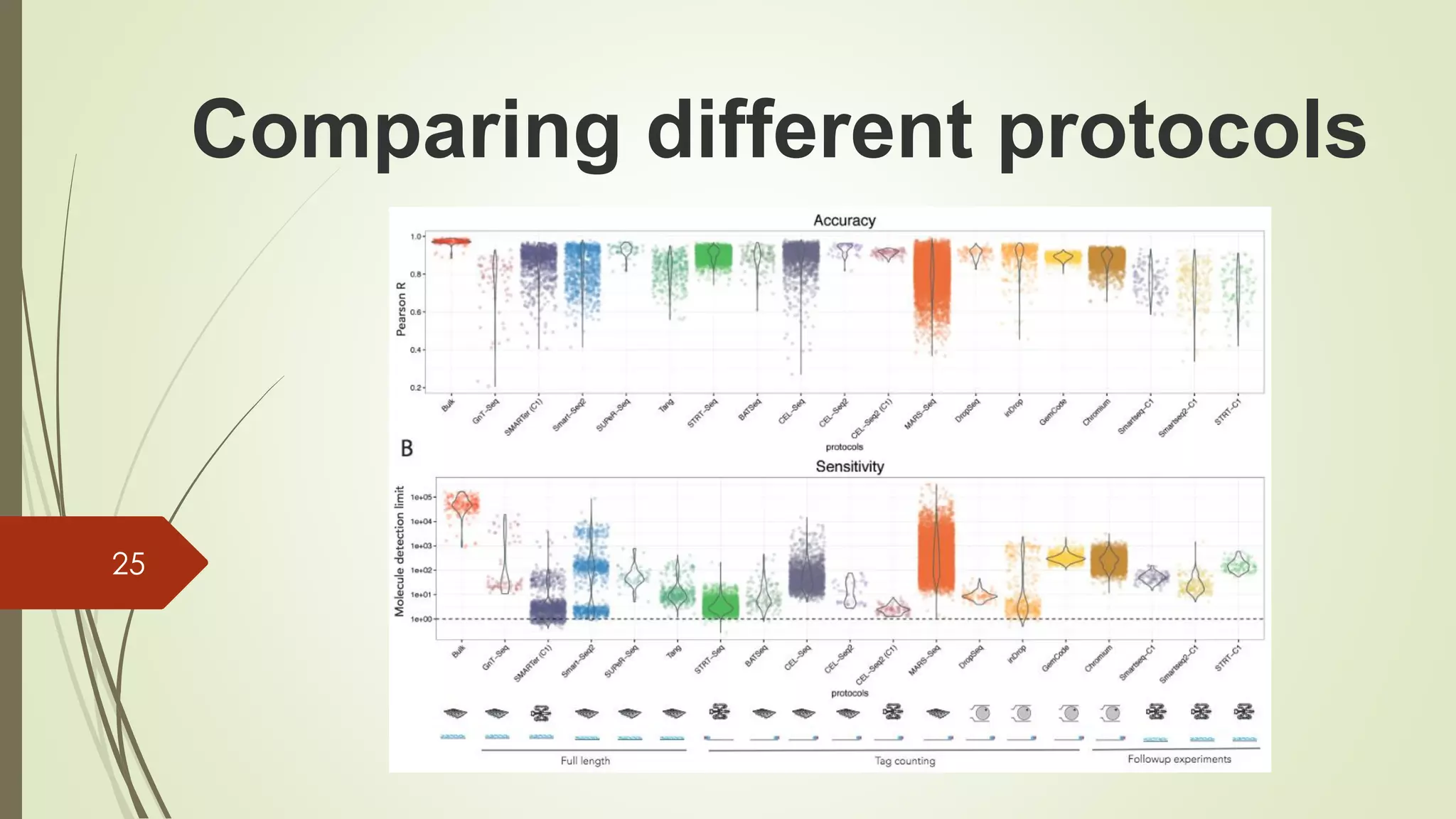
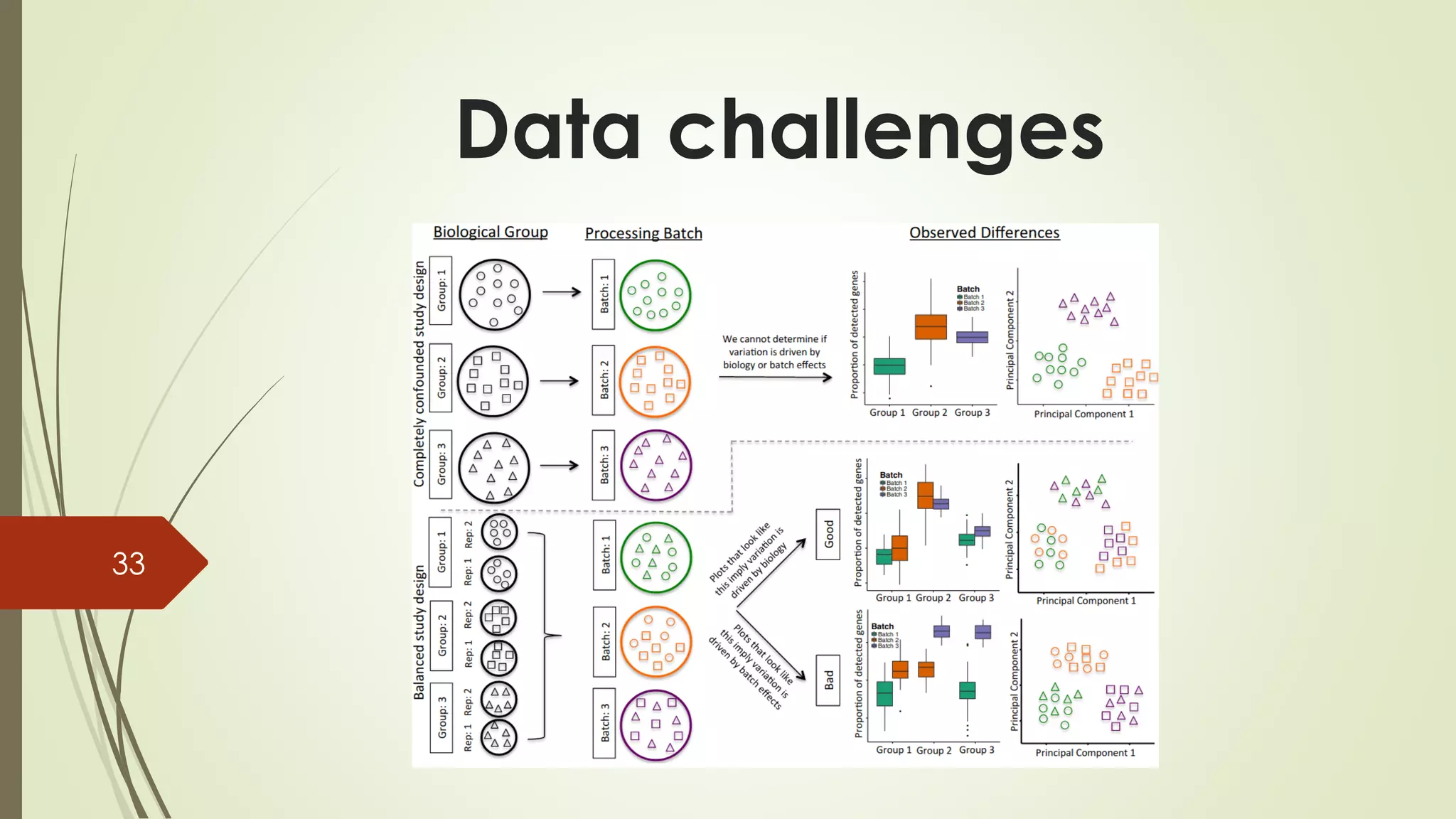
The document provides an overview of single-cell RNA sequencing (scRNA-seq), highlighting its advantages over bulk RNA sequencing, such as the ability to profile individual cell transcriptomes and study heterogeneous tissues. It discusses sample preparation protocols, including cell capture methods and transcript quantification techniques, as well as considerations for experimental design to avoid biases. Additionally, it compares different protocols and their sensitivities, emphasizing the importance of selecting the appropriate method based on research goals.