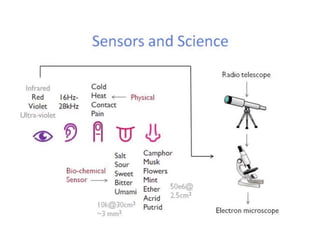
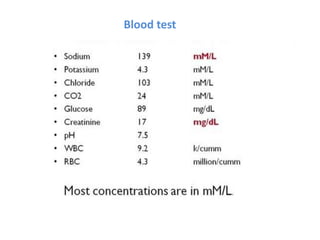
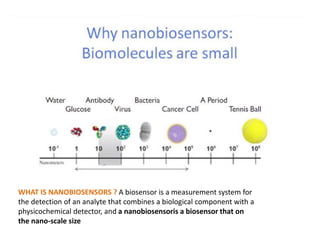
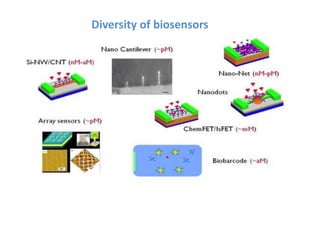
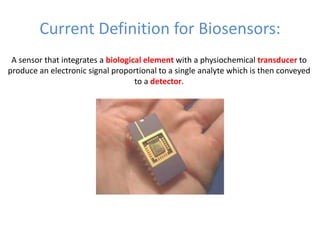
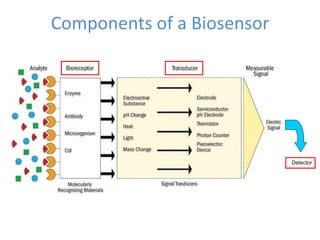
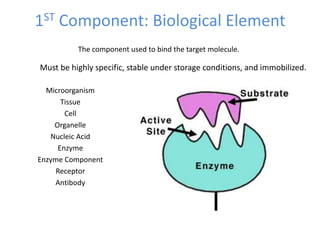
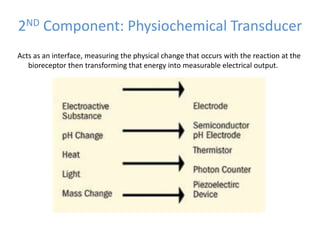
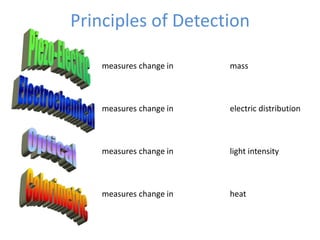
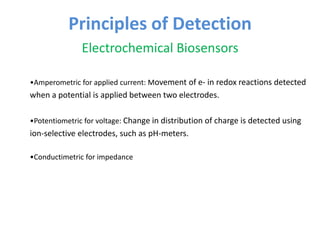
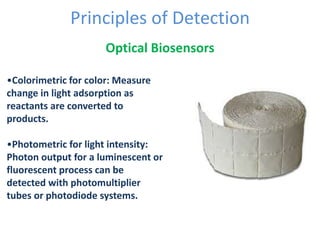
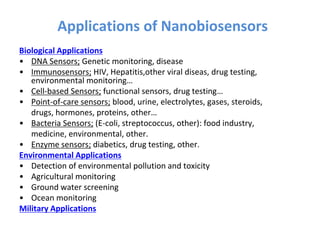
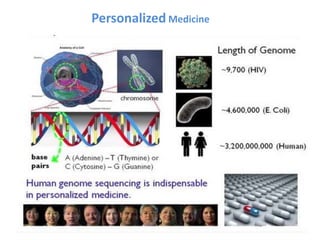
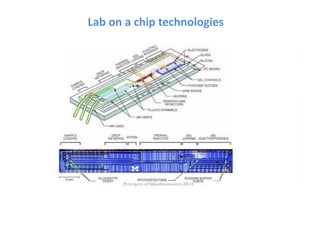
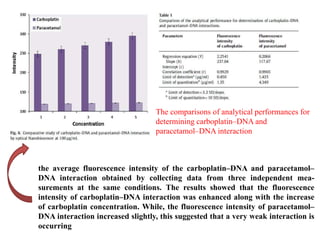
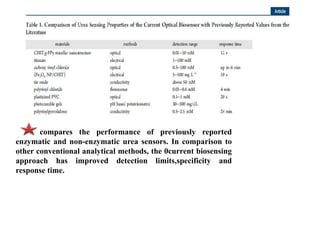
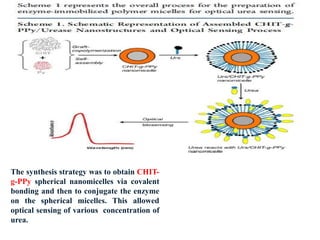
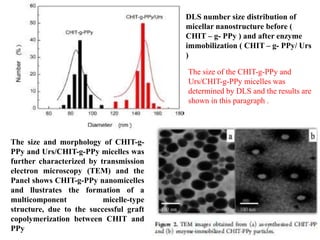
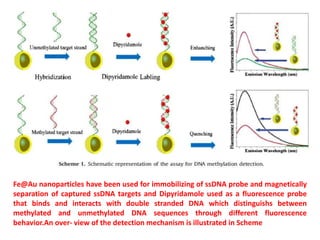
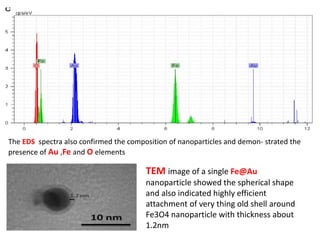
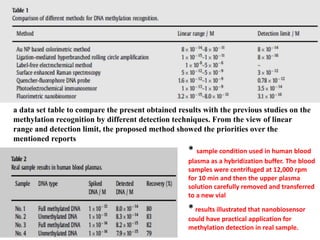
The document discusses nanobiosensors, which are measurement systems that integrate biological elements with physicochemical detectors at the nanoscale for high sensitivity in analyte detection. It details various components and detection principles of biosensors, including electrochemical, optical, and calorimetric methods, along with their applications in biological and environmental monitoring. Specific examples include the development of optical nanobiosensors for monitoring drug-DNA interactions and enhanced sensitivity in detecting DNA methylation in human plasma.