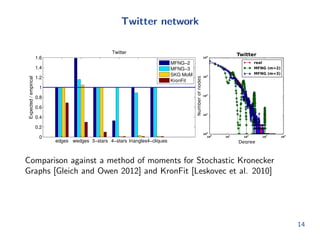
The document presents methods for modeling large networks using multifractal network generators (MFNG), focusing on scalability and estimation. It outlines a theoretical framework that allows for efficient graph generation and parameter fitting through method of moments estimation, demonstrating effectiveness with empirical data from synthetic and real-world networks. The authors also discuss challenges and solutions for fast sampling and estimating network structures.

![Setting
We want a simple, scalable method to model networks and generate
random (undirected) graphs
I Looking for graph generators that can mimic real world graph
structure
{ Power law degree distribution,
{ High clustering coecient, etc.
I Many models have been proposed, starting with Erdos-Renyi graphs
I Relatively recent models: SKG [Leskovec et al. 2010], BTER
[Seshadhri et al. 2012], TCL [Pfeier et al. 2012]
I In 2011 Palla et al. introduce multifractal network generators,
`generalizing' SKG
2](https://image.slidesharecdn.com/multifractal-kdd-2014-140906183143-phpapp02/85/Learning-multifractal-structure-in-large-networks-KDD-2014-2-320.jpg)


![ed lenghts li
I Number of recursive levels: k logm(n)
I Probabilities of edges between nodes, based on categories, stored in
matrix P 2 [0; 1]mm
4](https://image.slidesharecdn.com/multifractal-kdd-2014-140906183143-phpapp02/85/Learning-multifractal-structure-in-large-networks-KDD-2014-7-320.jpg)
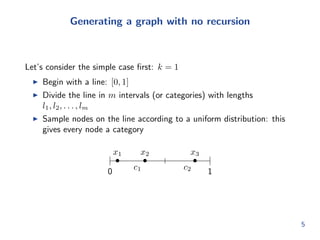
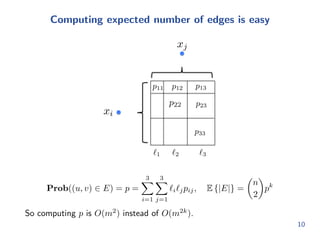
![rst: k = 1
I Begin with a line: [0; 1]
I Divide the line in m intervals (or categories) with lengths
l1; l2; : : : ; lm
I Sample nodes on the line according to a uniform distribution: this
gives every node a category
x2
x3
c1 c2
x1
0 1
5](https://image.slidesharecdn.com/multifractal-kdd-2014-140906183143-phpapp02/85/Learning-multifractal-structure-in-large-networks-KDD-2014-9-320.jpg)

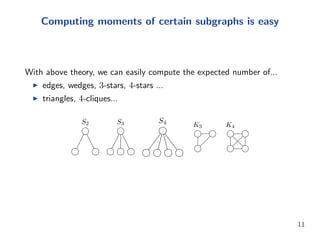
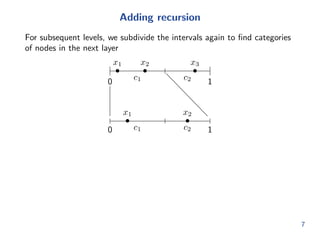
![Expanding the recursion
So we can get a full probabilistic adjacency matrix Q 2 [0; 1]mkmk
by
expanding all recursive levels
Problem: Q grows fast with k. Dicult to do inference.
Intuitively, we should not have to do this.
8](https://image.slidesharecdn.com/multifractal-kdd-2014-140906183143-phpapp02/85/Learning-multifractal-structure-in-large-networks-KDD-2014-15-320.jpg)
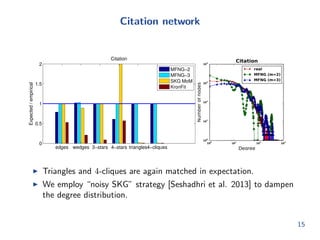
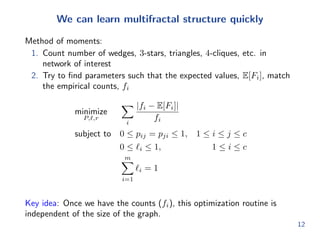
![nd parameters such that the expected values, E[Fi], match
the empirical counts, fi
minimize
P;`;r
X
i
jfi E[Fi]j
fi
subject to 0 pij = pji 1; 1 i j c
0 `i 1; 1 i c
Xm
i=1
`i = 1
Key idea: Once we have the counts (fi), this optimization routine is
independent of the size of the graph.
12](https://image.slidesharecdn.com/multifractal-kdd-2014-140906183143-phpapp02/85/Learning-multifractal-structure-in-large-networks-KDD-2014-20-320.jpg)