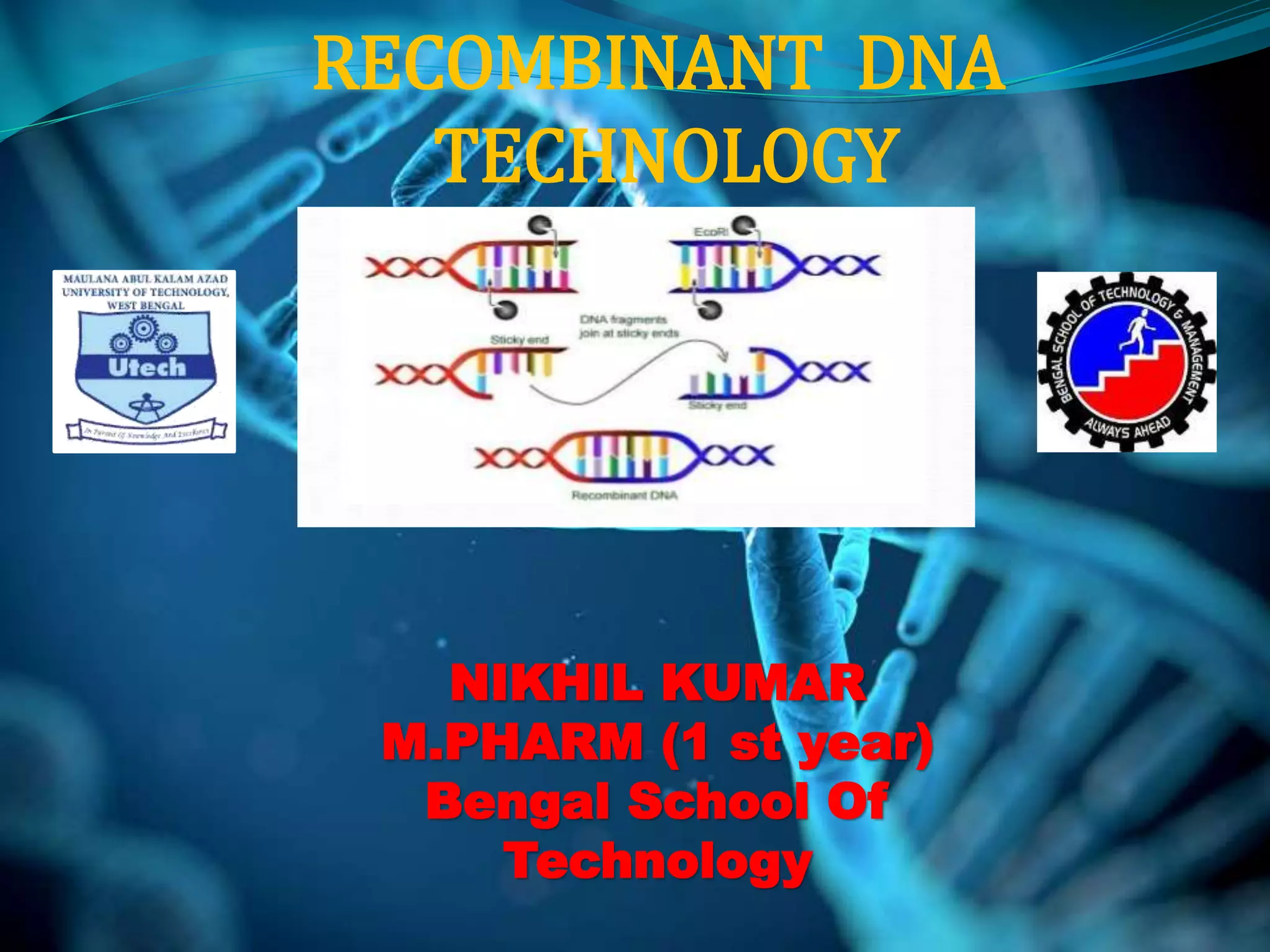
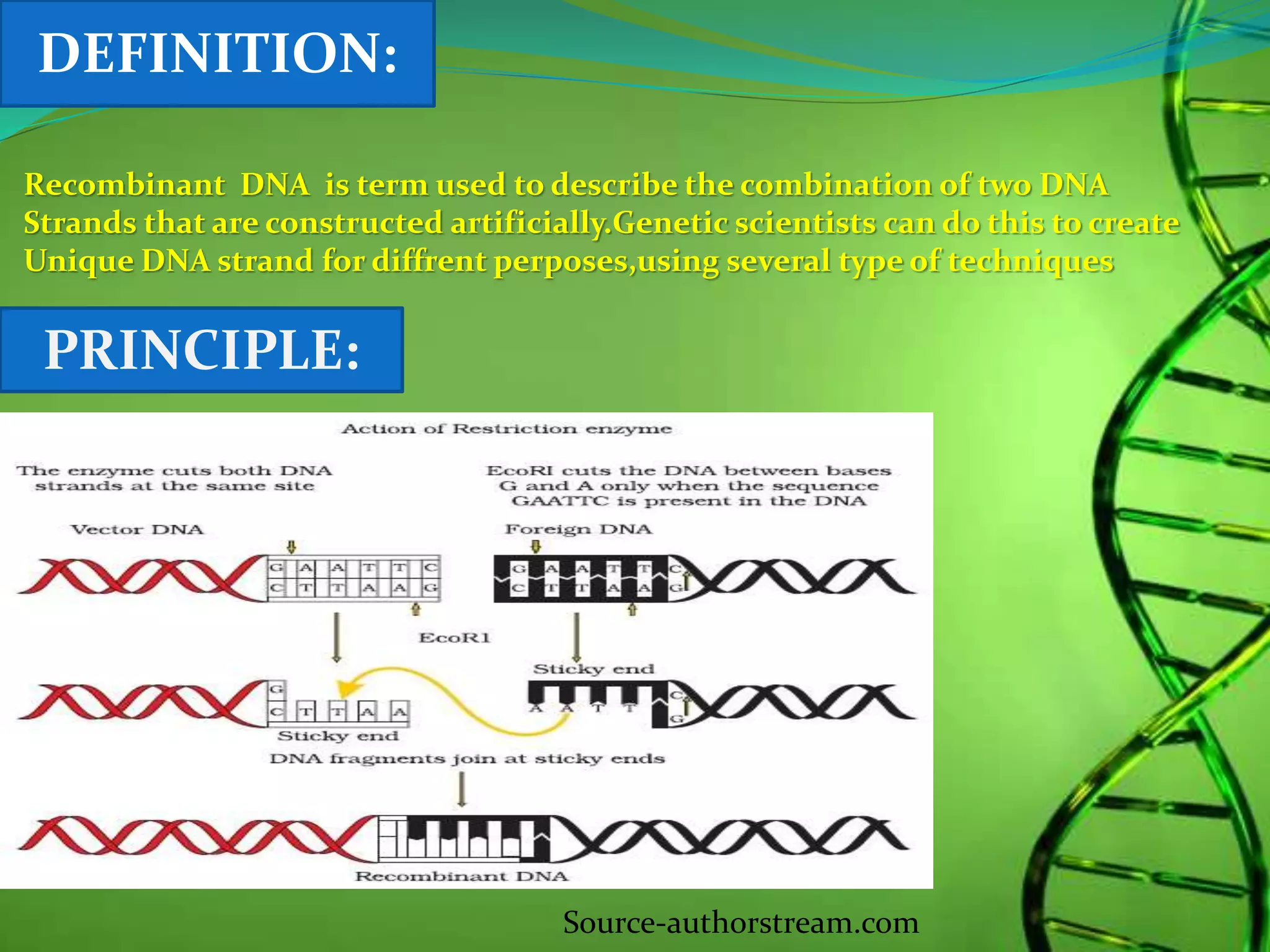
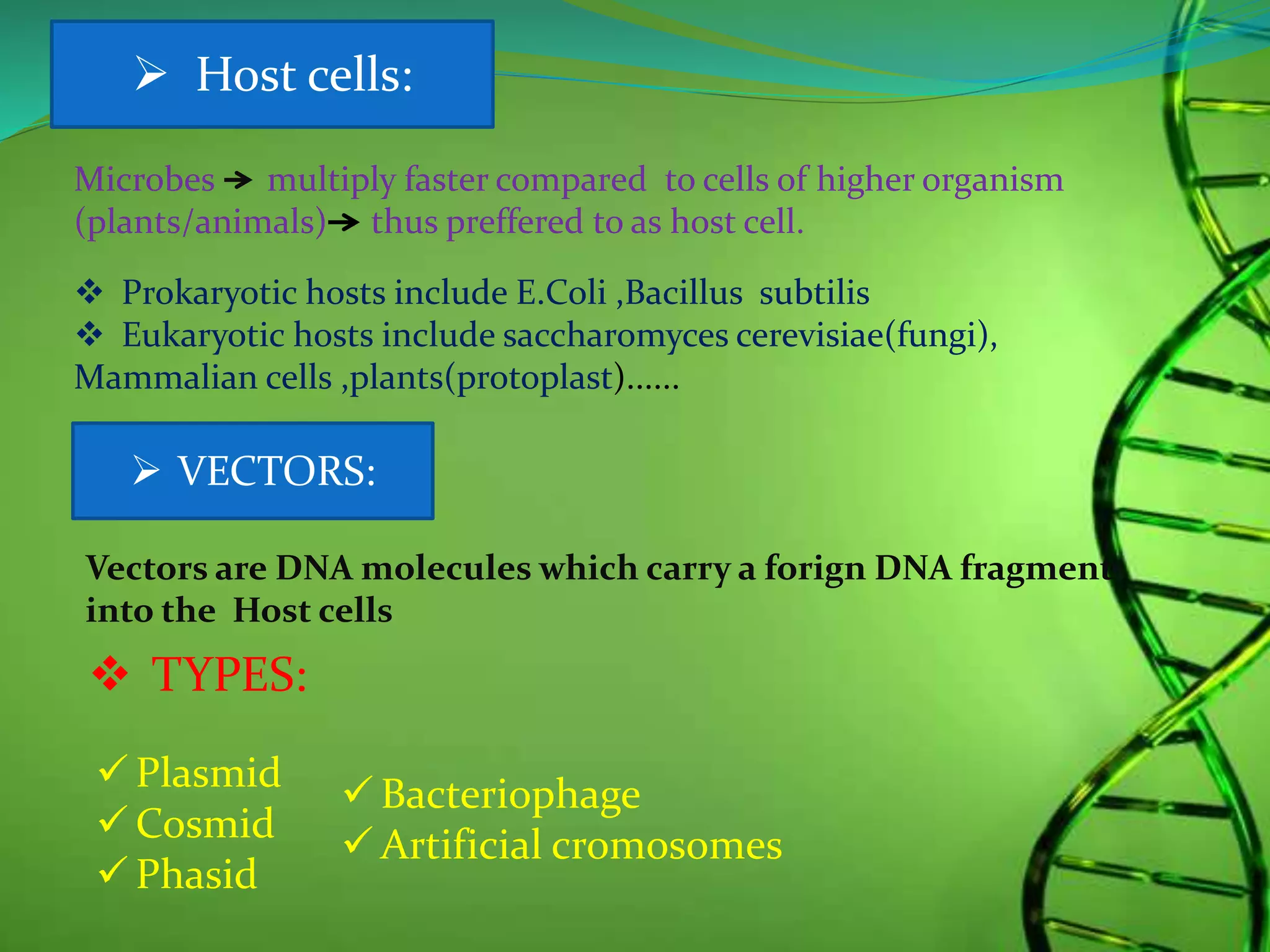
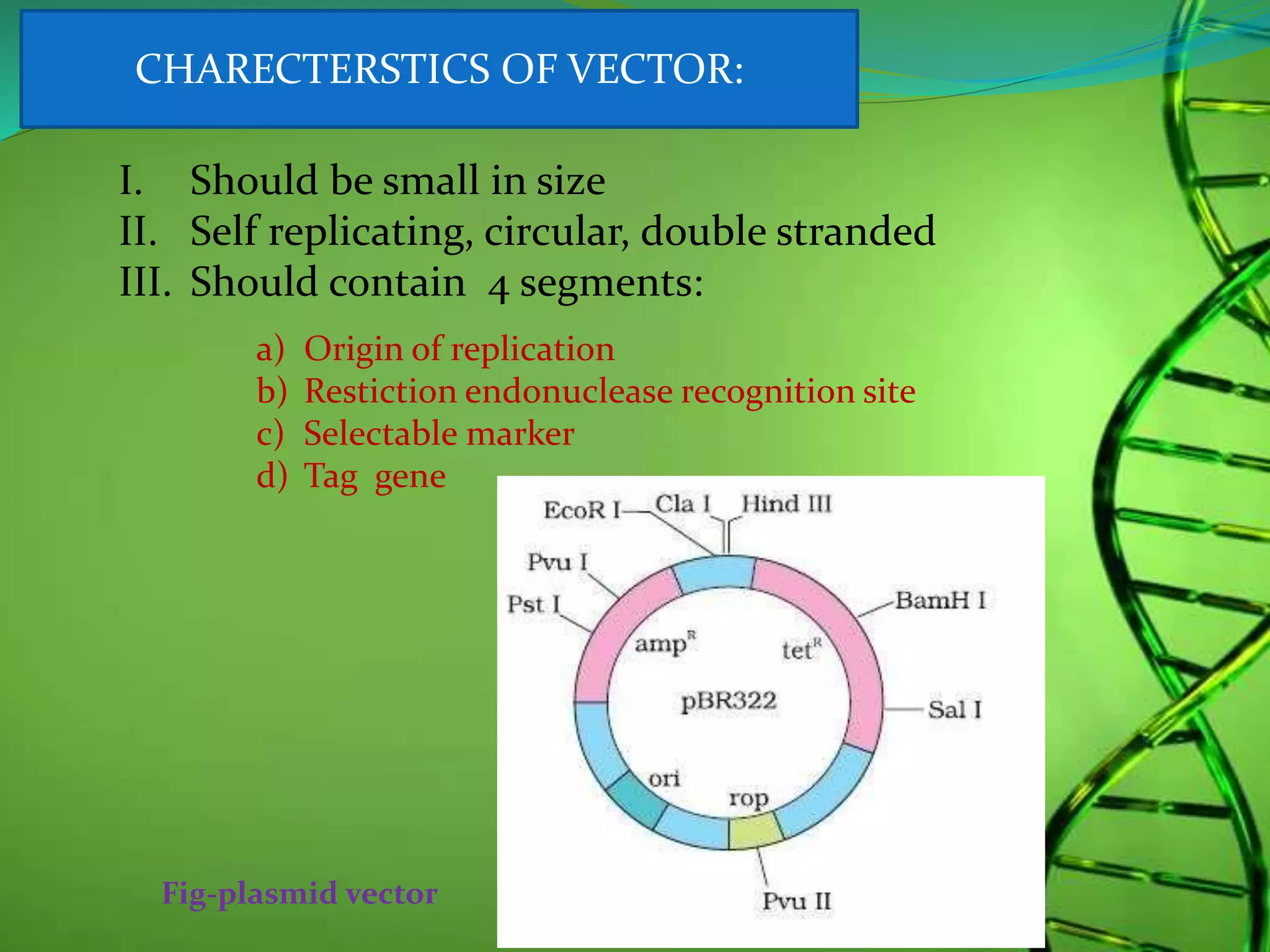
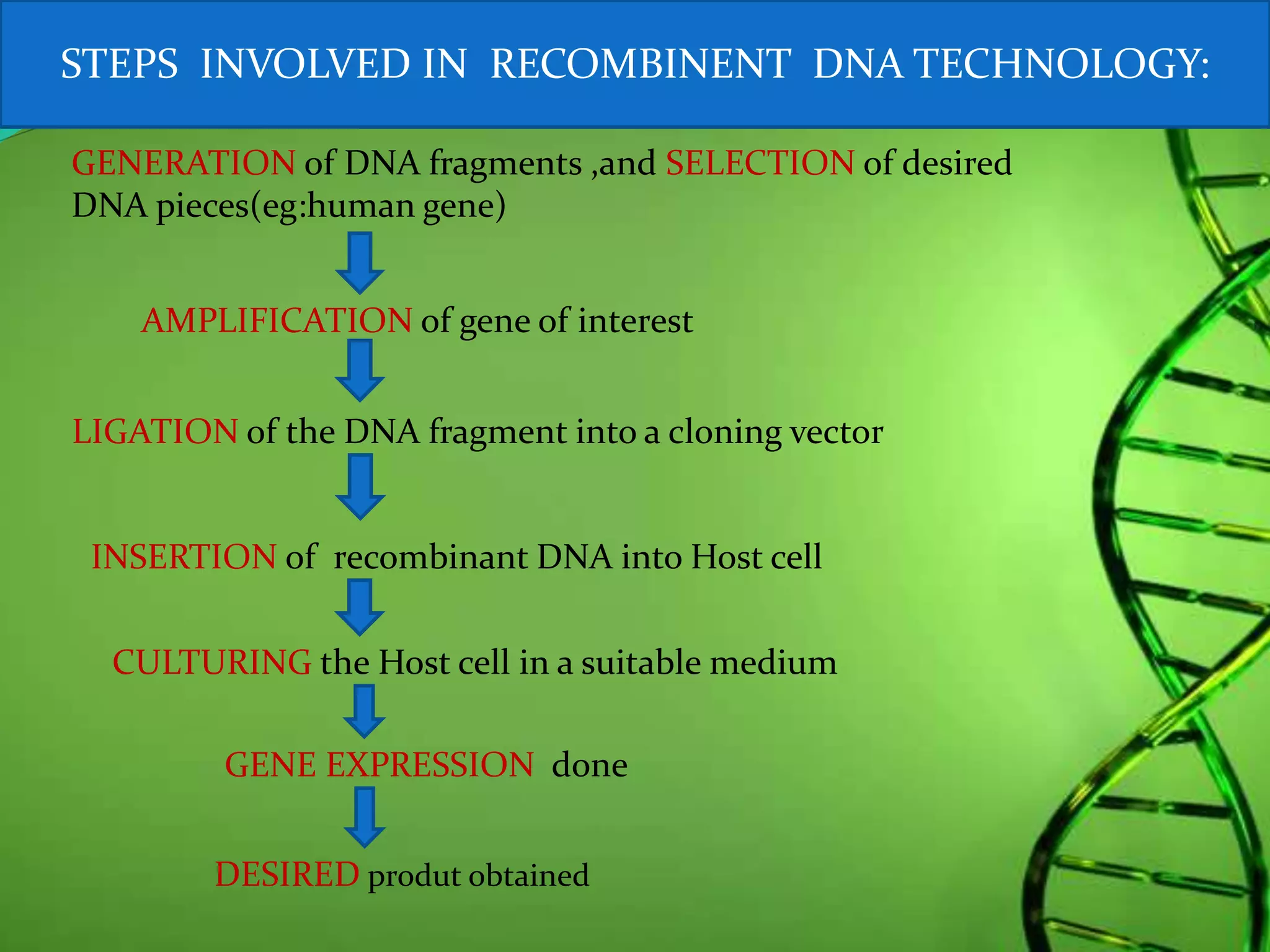
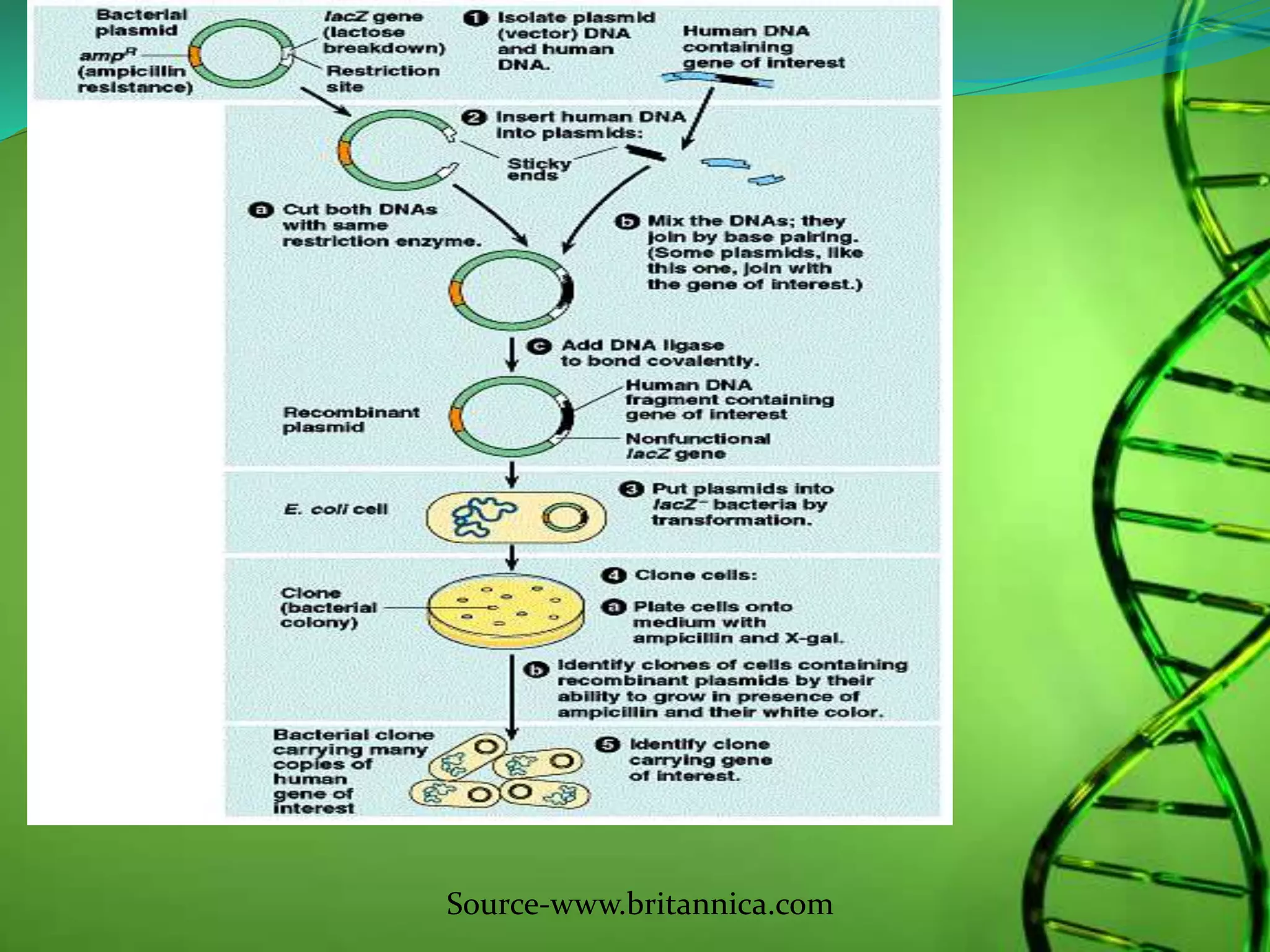
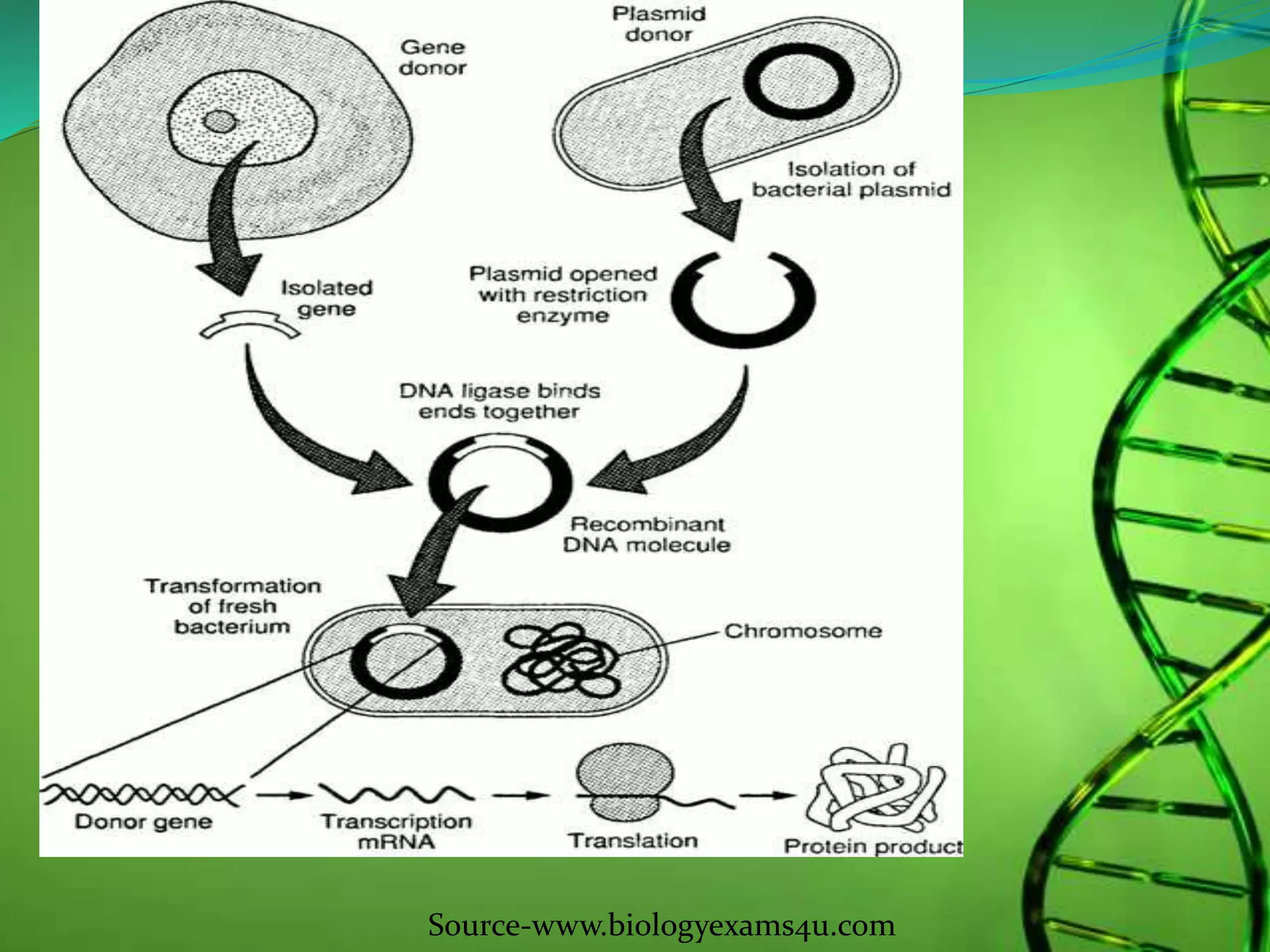
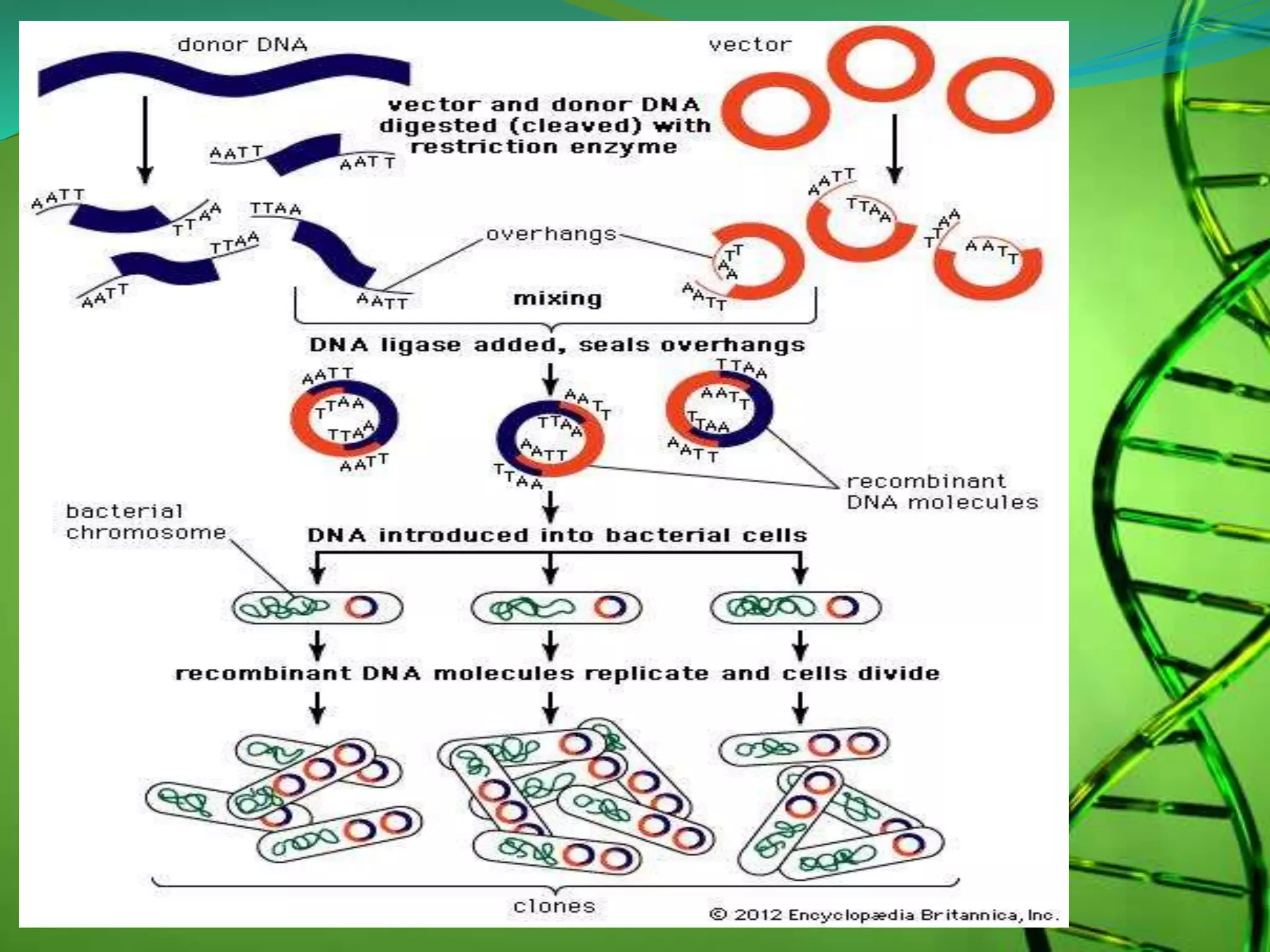
Recombinant DNA technology involves combining DNA strands from different sources artificially. It uses restriction enzymes to cut DNA at specific sites, ligases to join DNA fragments, and vectors and host cells to replicate the recombinant DNA. The first successful recombinant DNA experiment joined DNA from Salmonella typhimurium. Key tools include restriction enzymes, ligases, vectors like plasmids, and host cells like E. coli. Applications include molecular diagnosis of diseases, monoclonal antibody production, gene therapy, and increasing crop productivity.