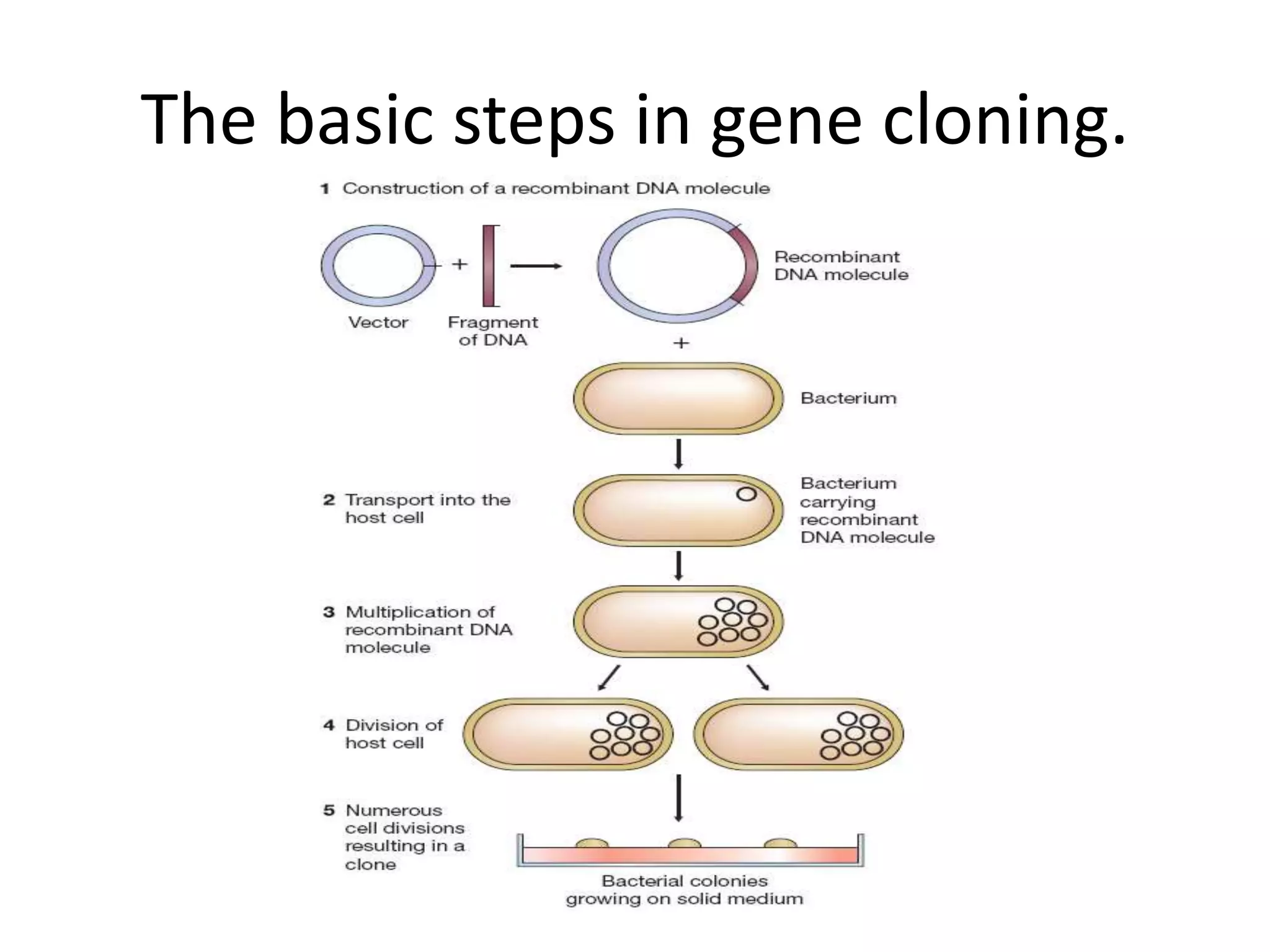
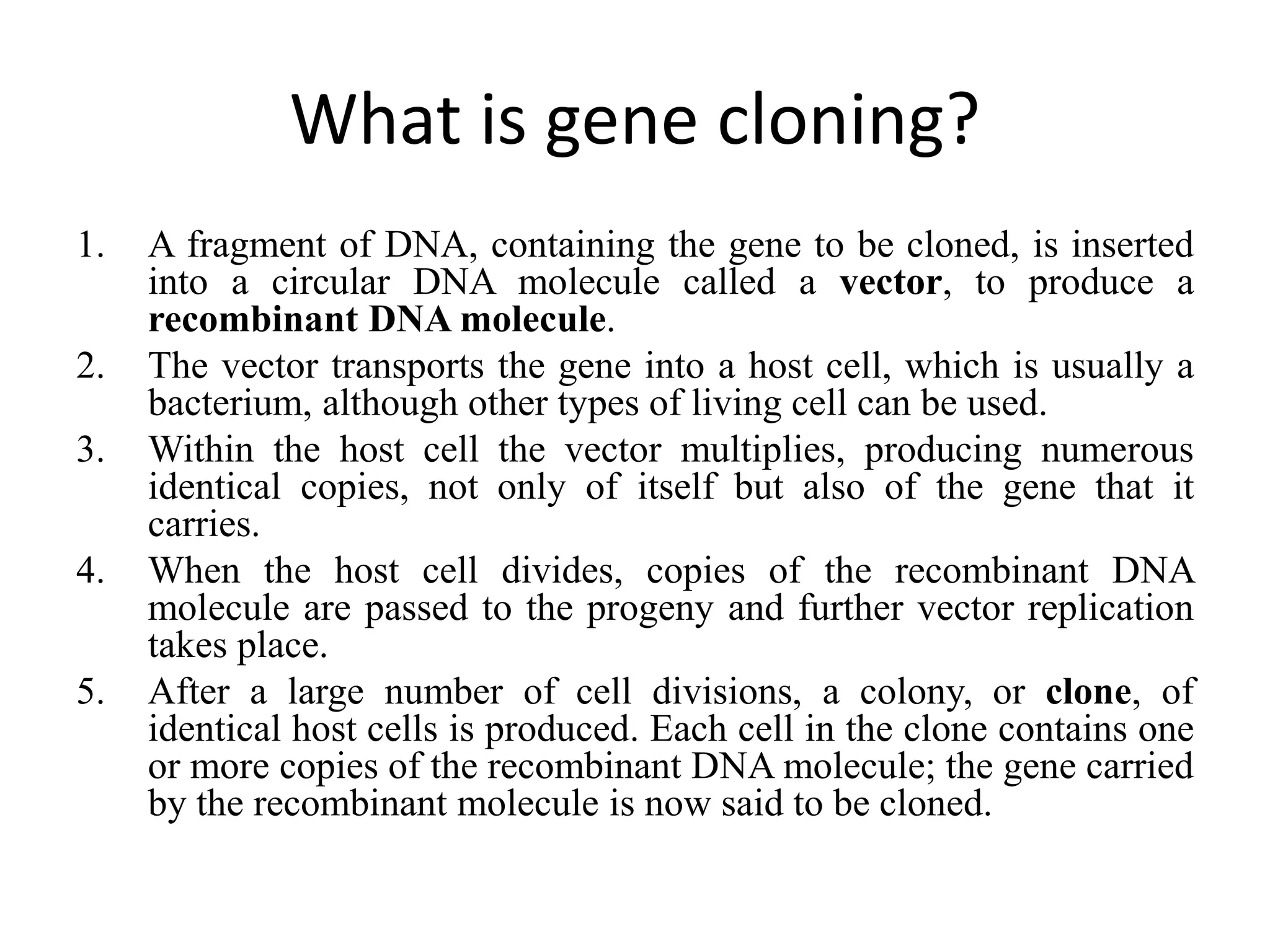
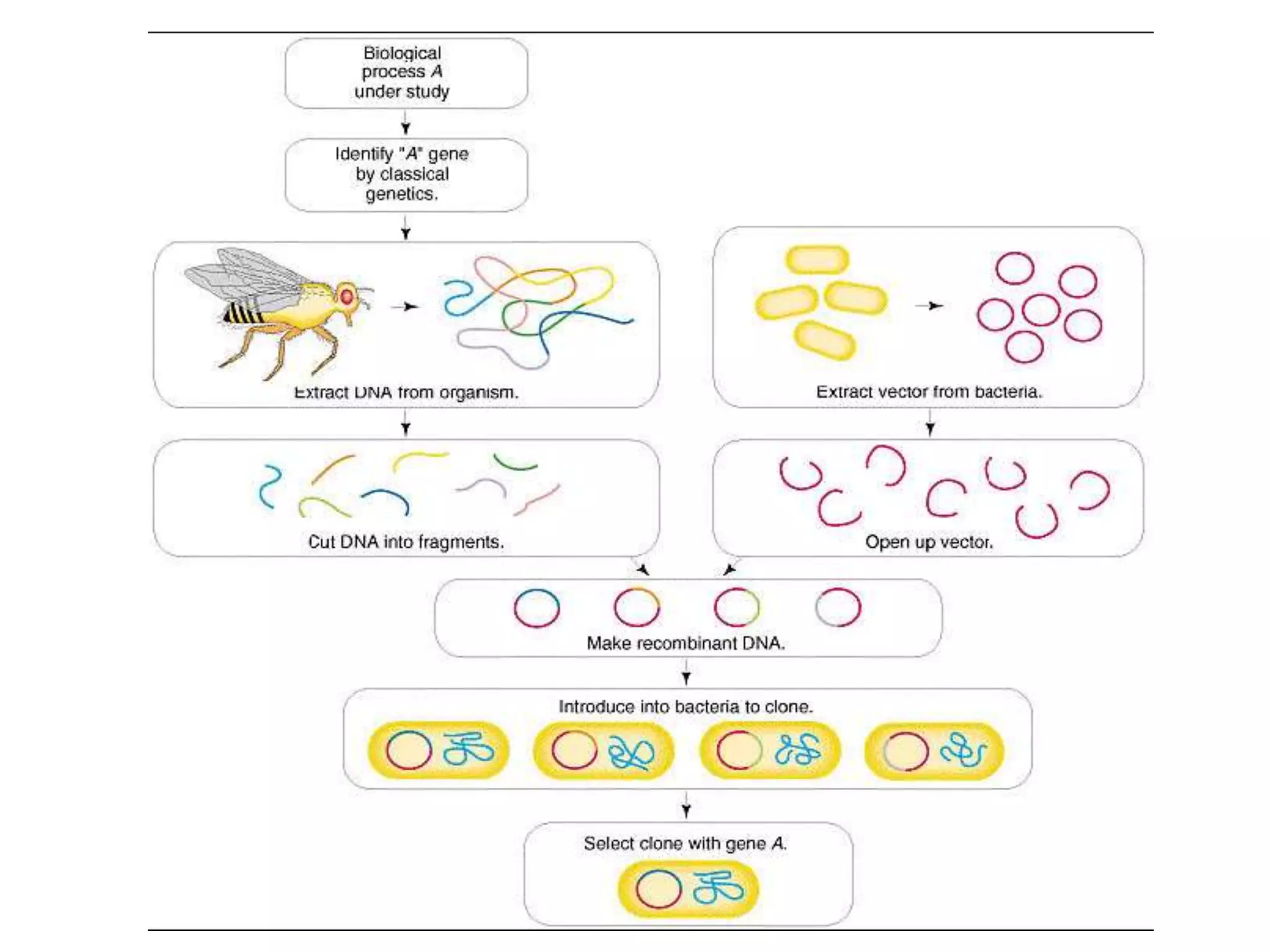
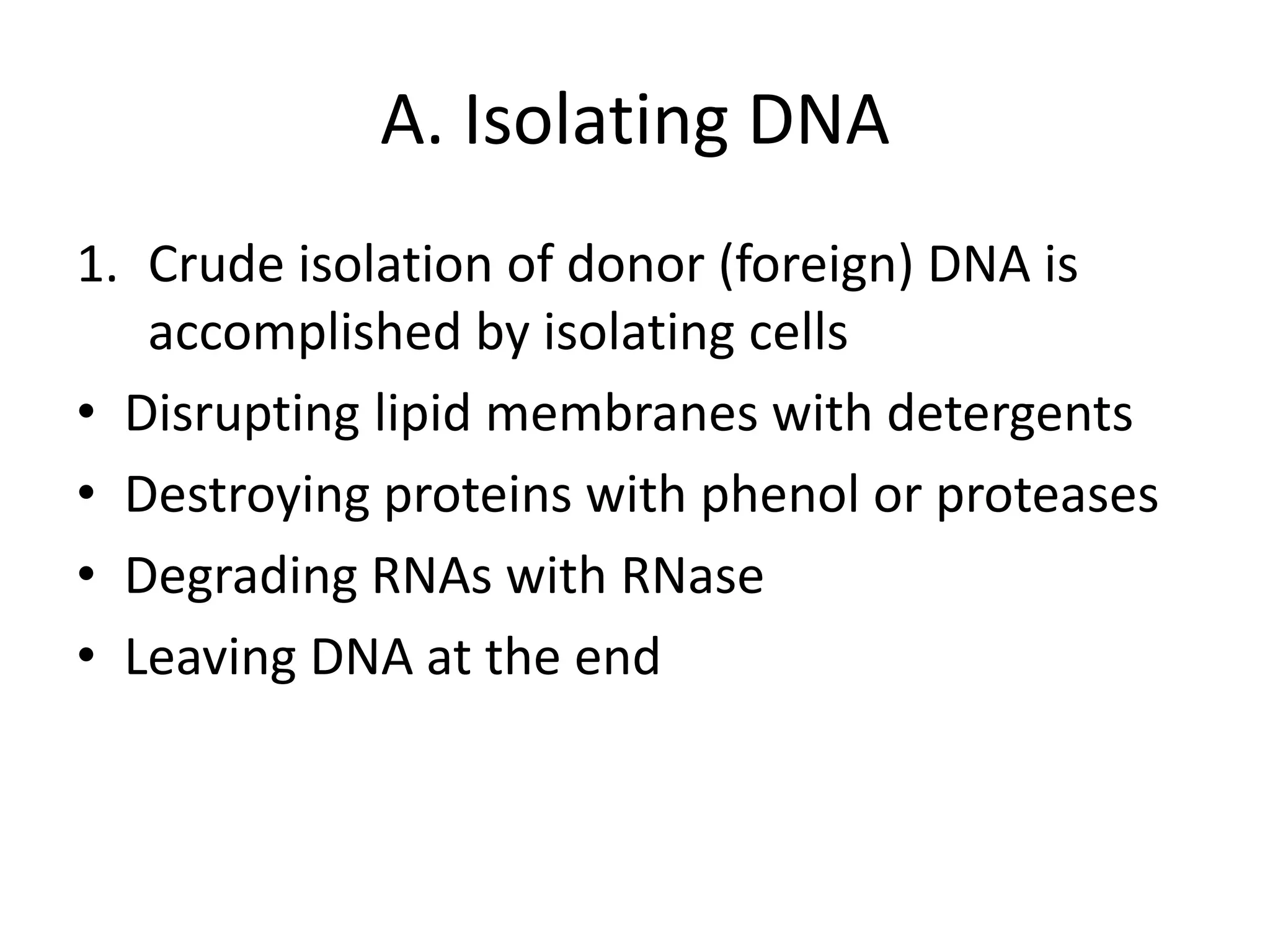
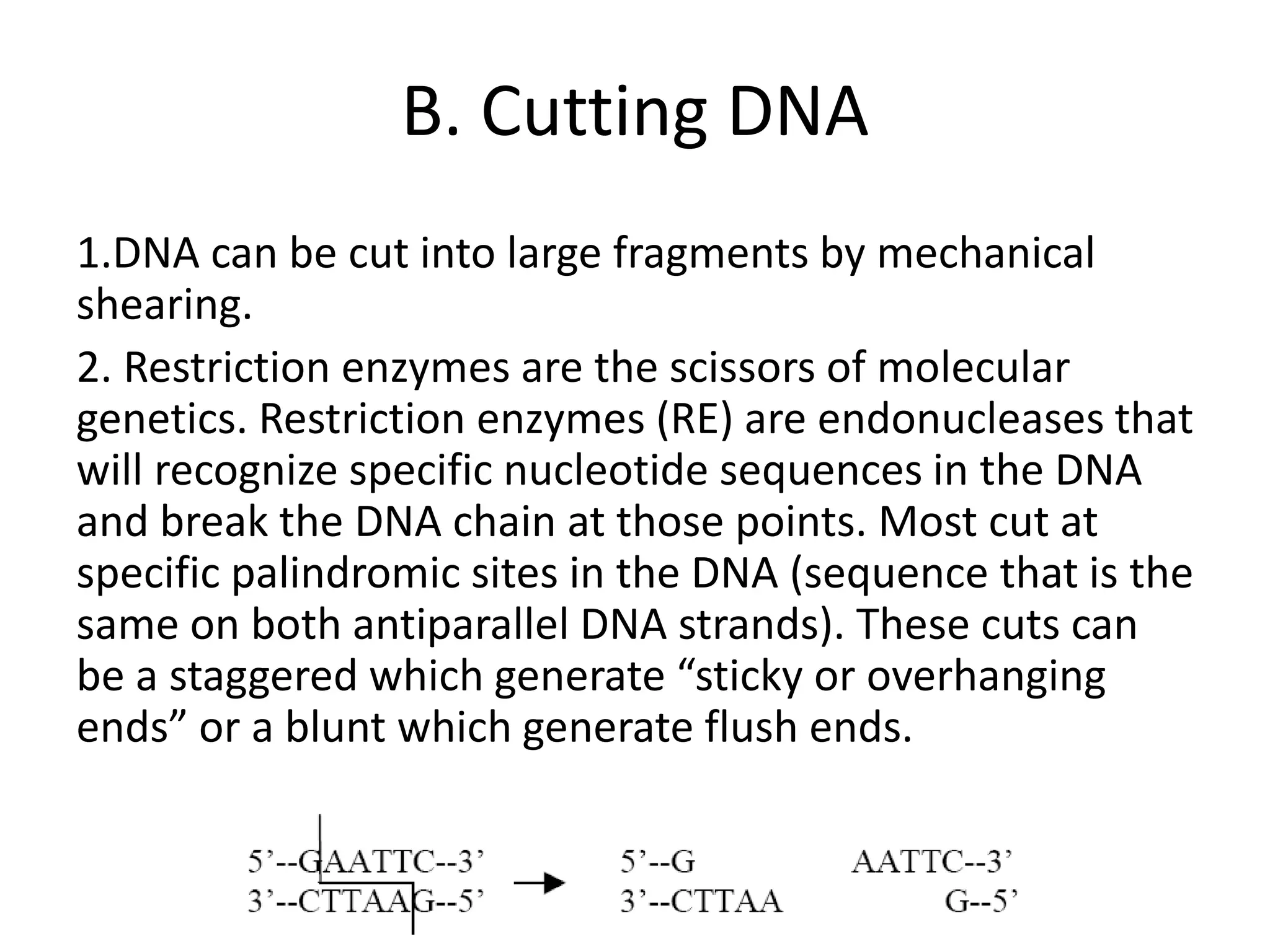
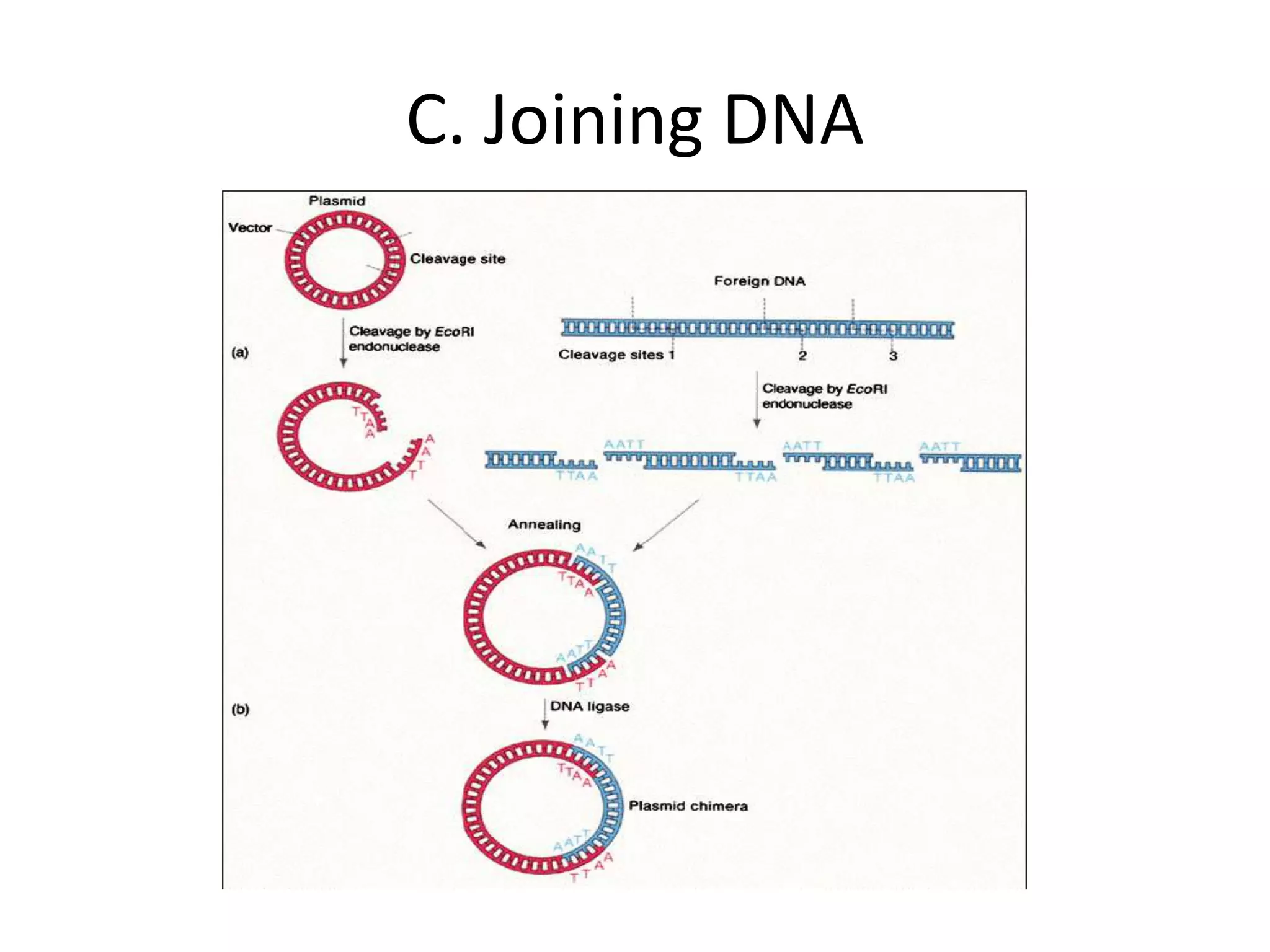
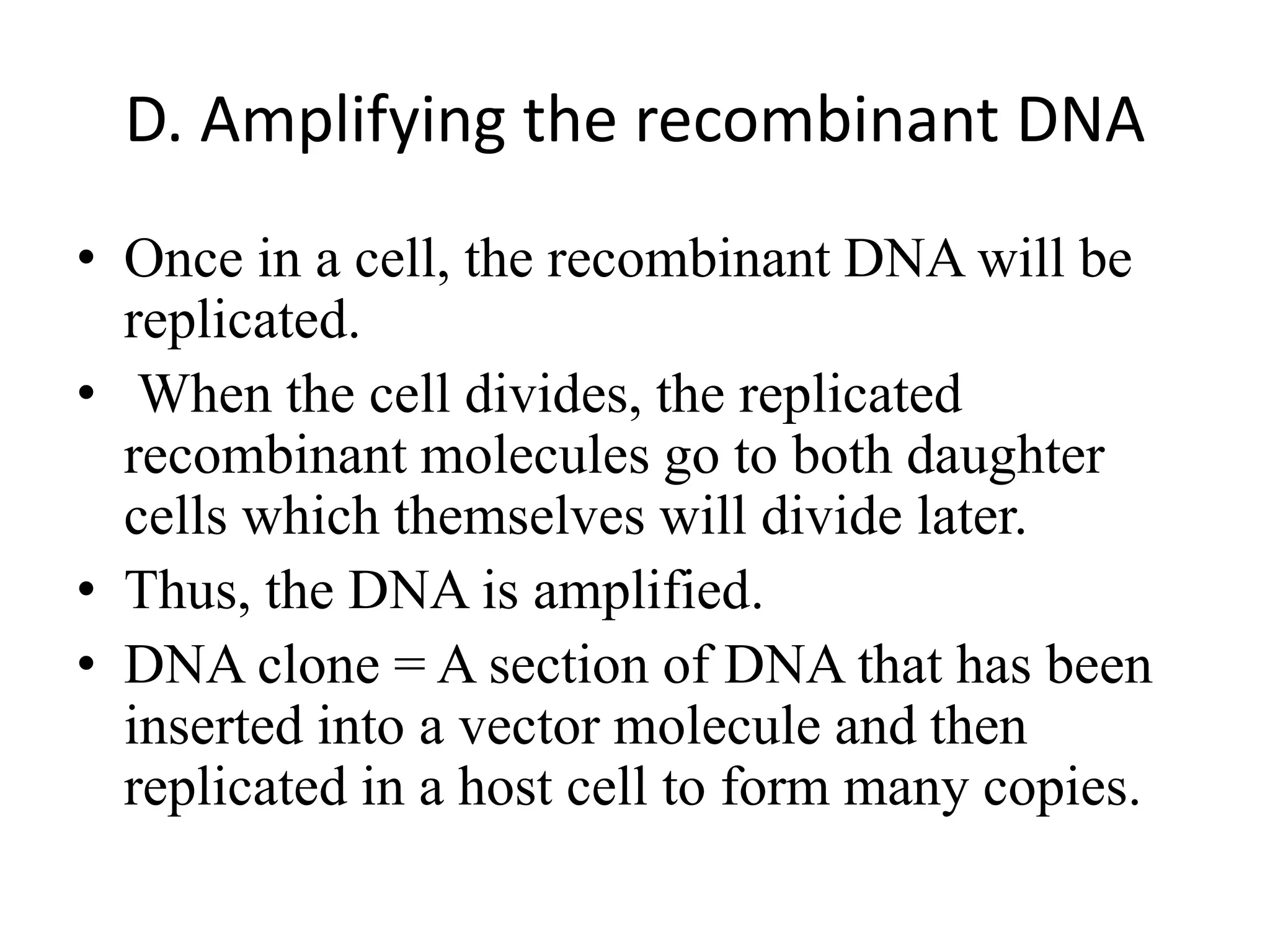
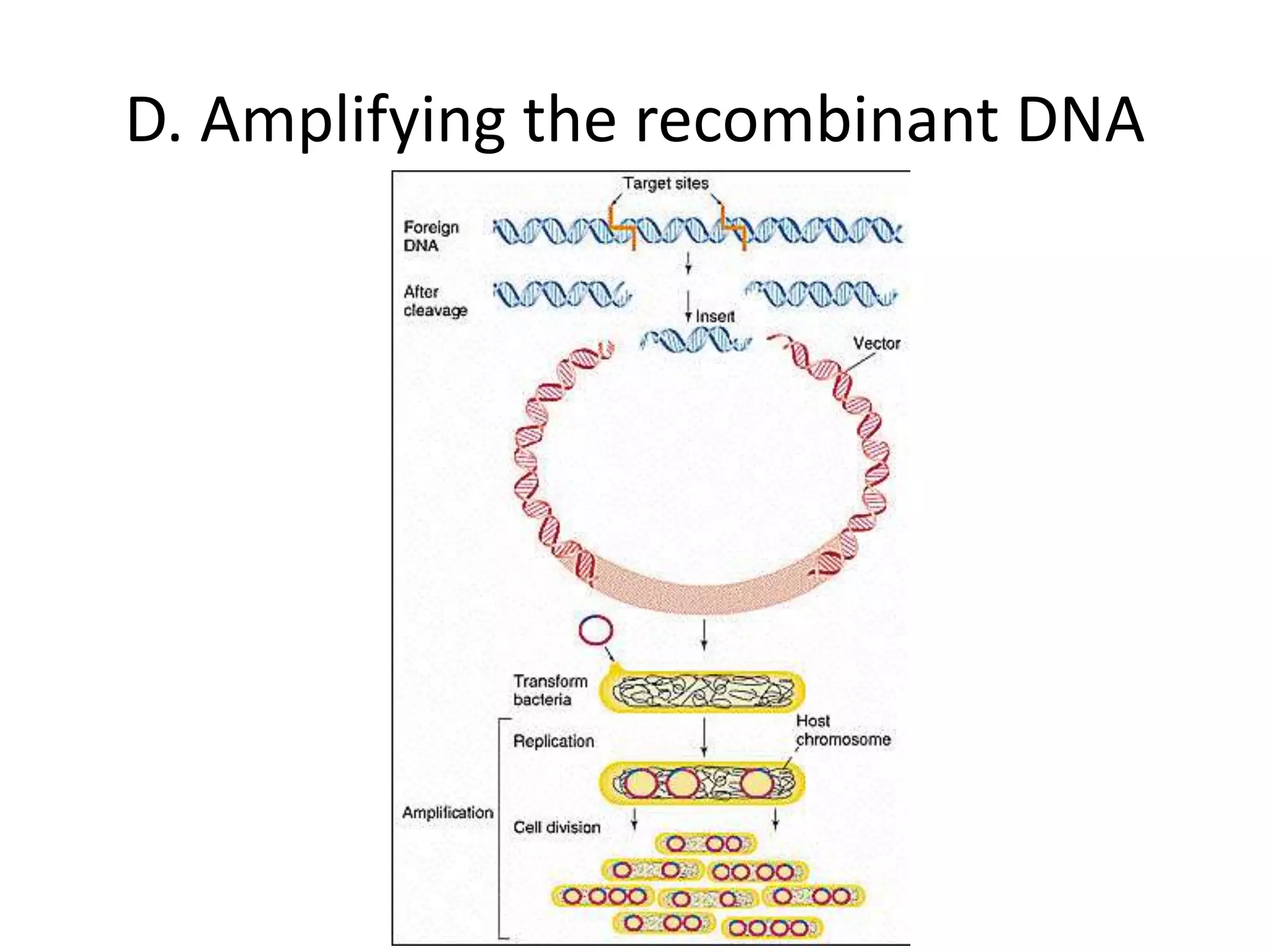
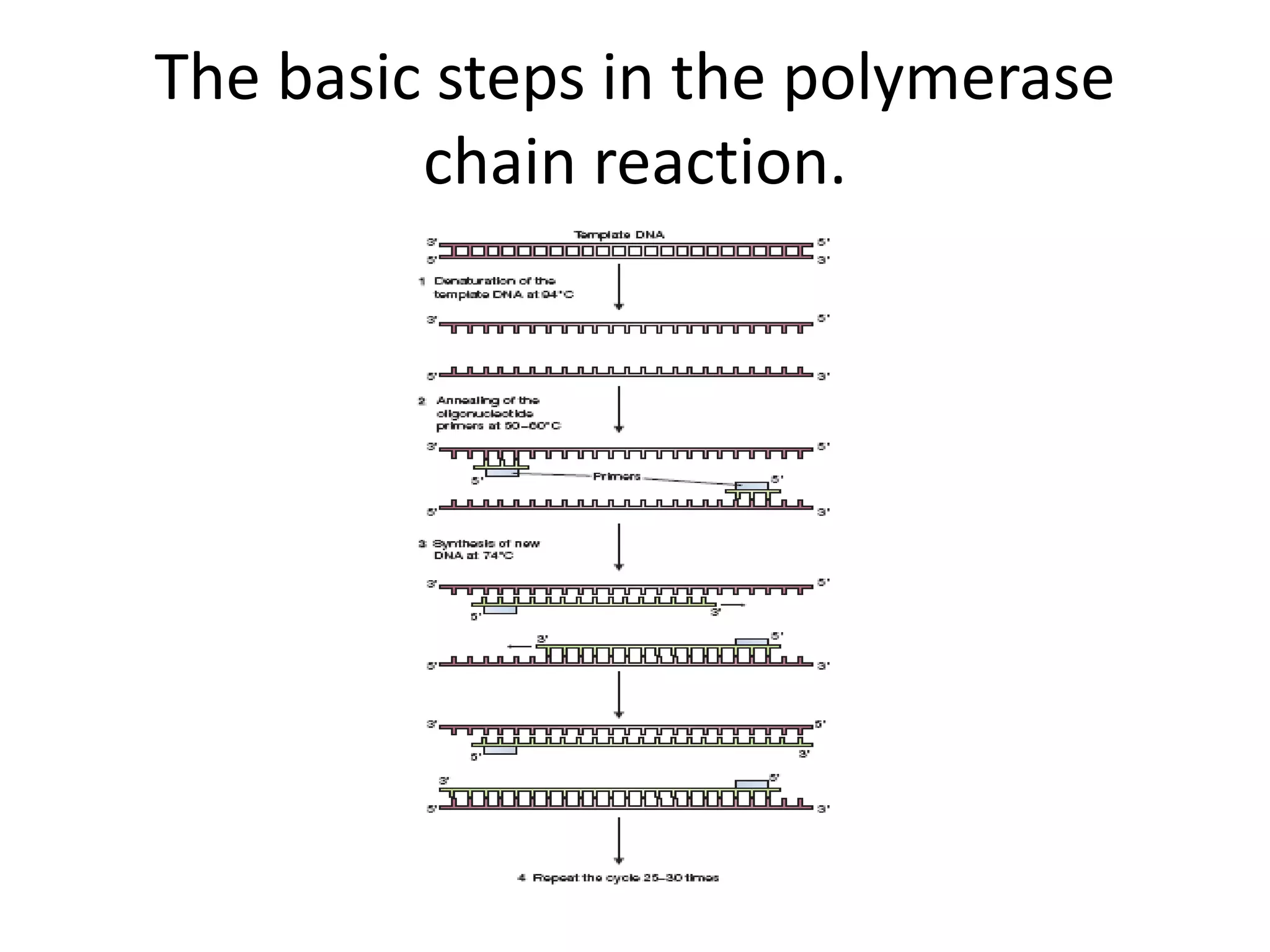
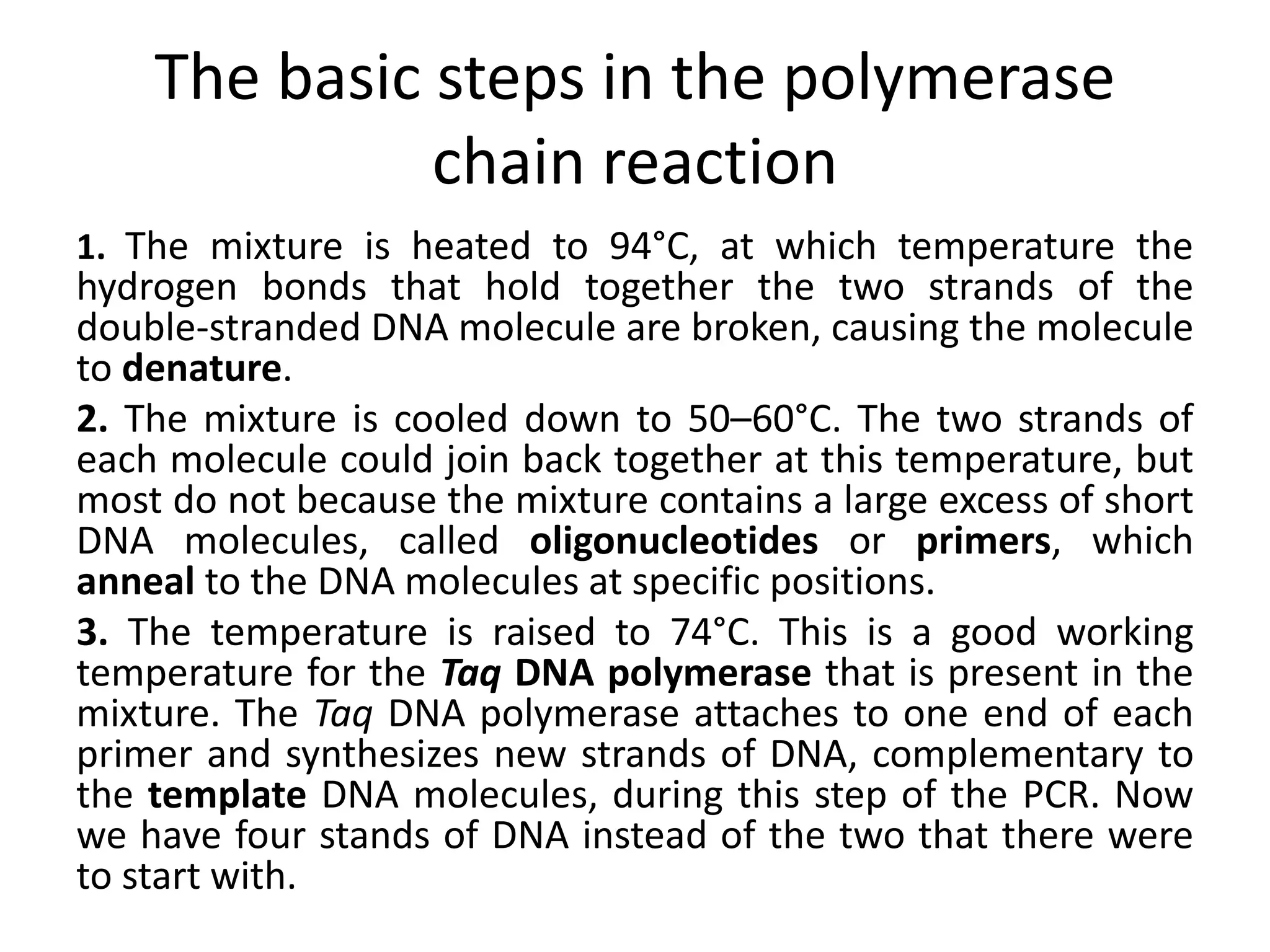
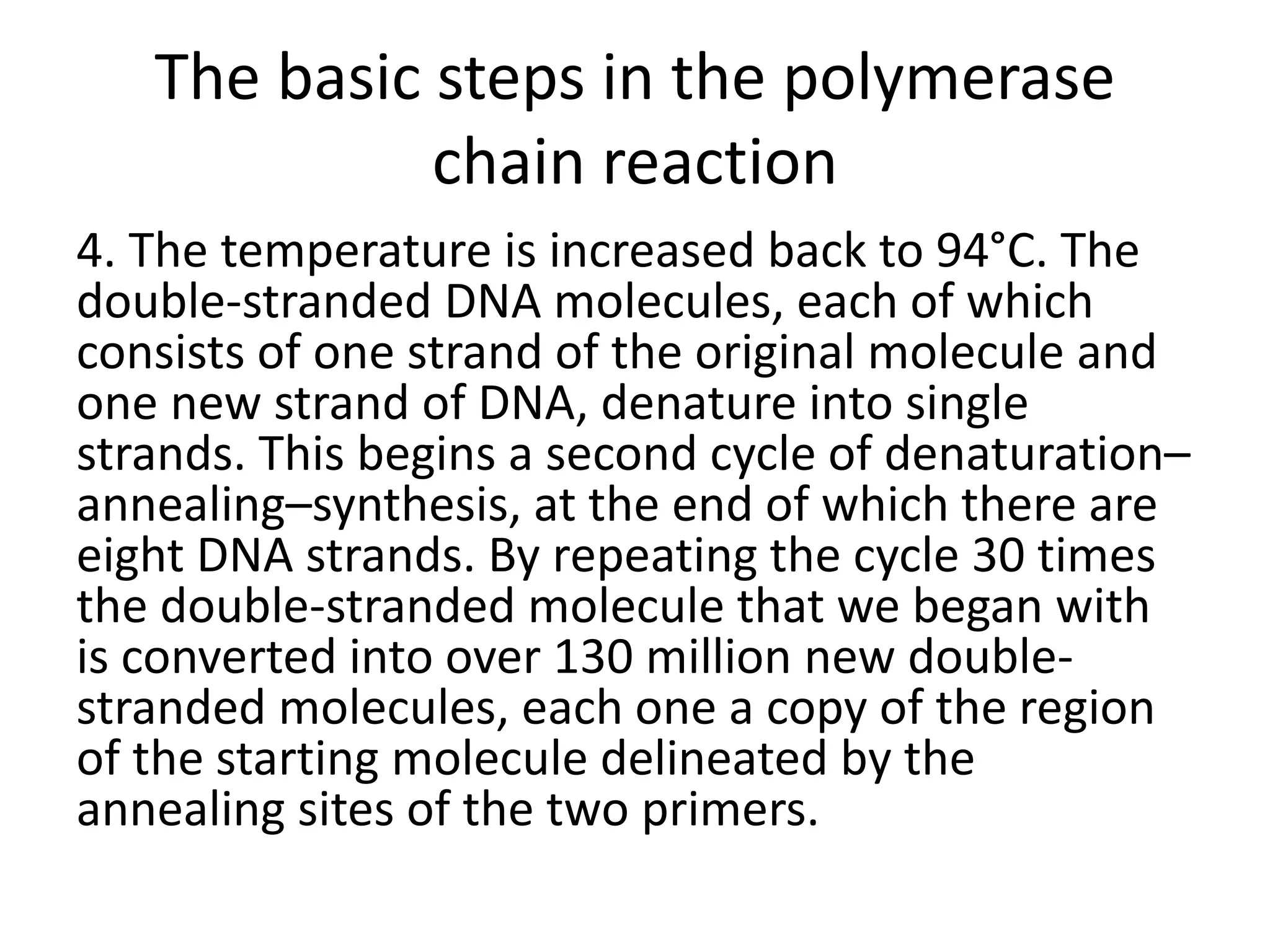
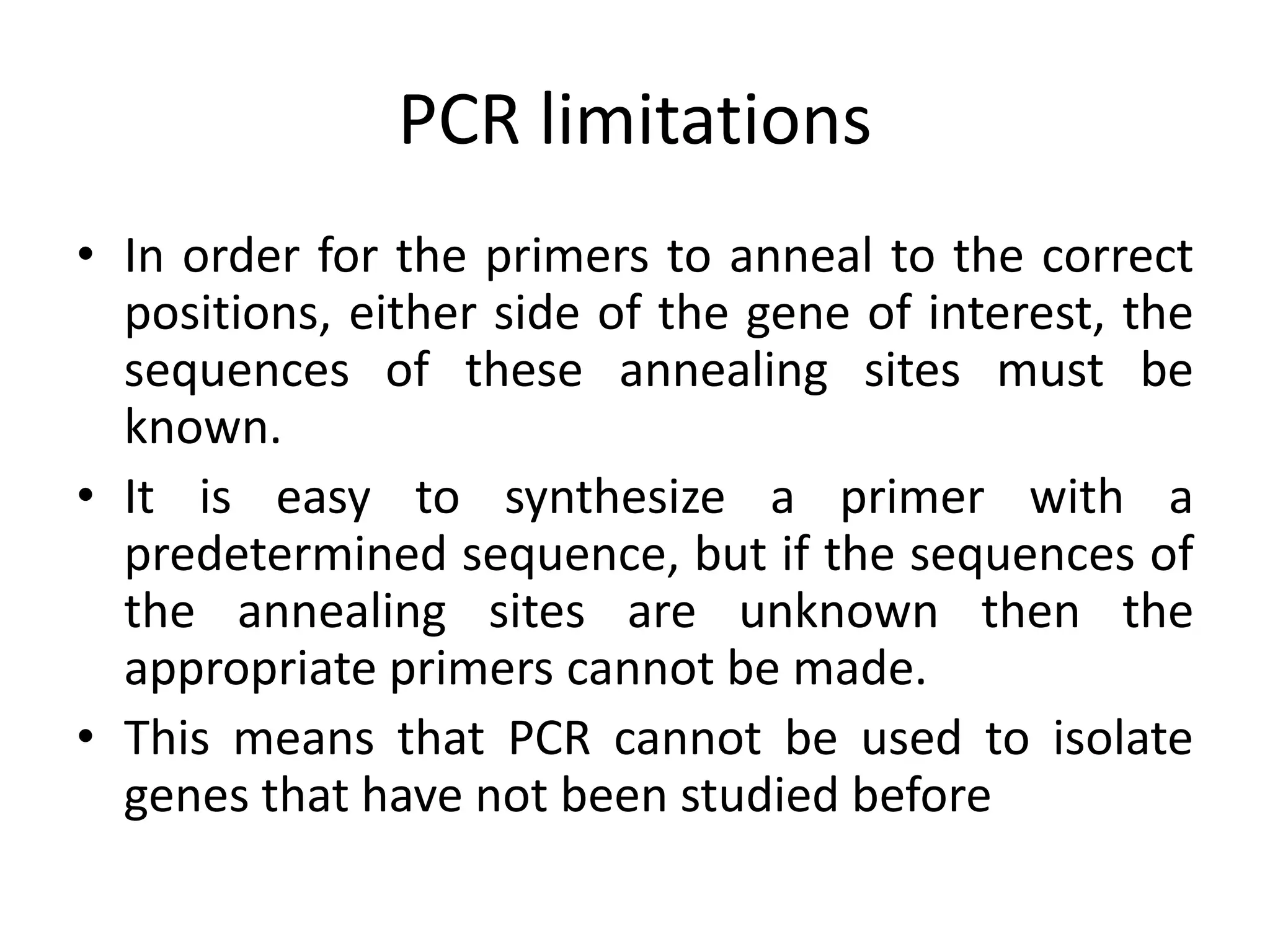
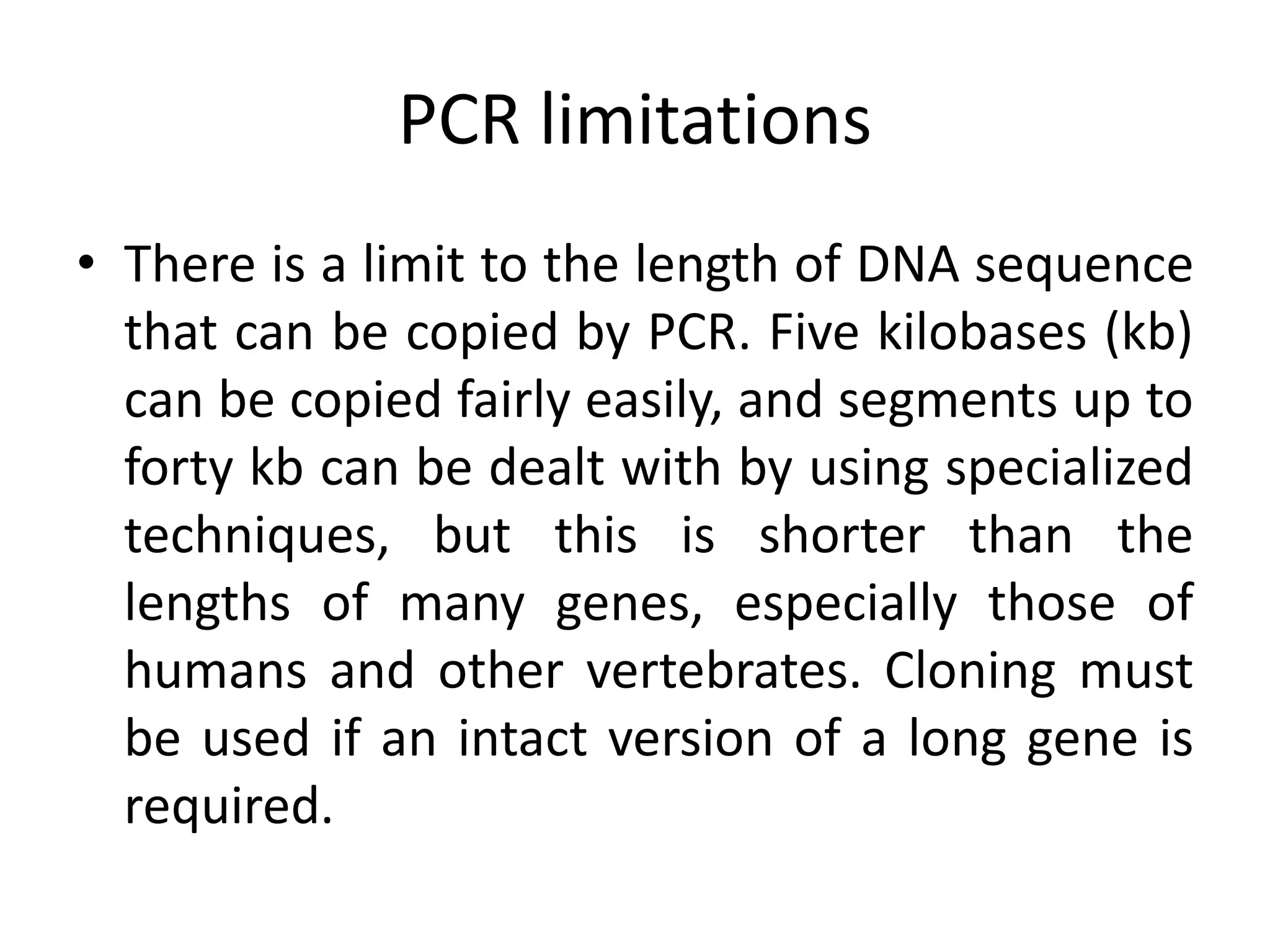
Recombinant DNA technology involves combining DNA from different sources and introducing it into a host cell. This allows for precise genetic analysis and practical applications. Key developments included elucidating DNA structure, cracking the genetic code, and describing transcription and translation. Gene cloning was developed in the 1970s, enabling previously impossible experiments. It involves isolating DNA, cutting it with restriction enzymes, ligating it into a vector, transforming host cells to amplify the recombinant DNA. The polymerase chain reaction (PCR) allows amplifying specific DNA regions without living cells by repeated heating and cooling in a test tube. It has revolutionized research fields like genetics and molecular biology.