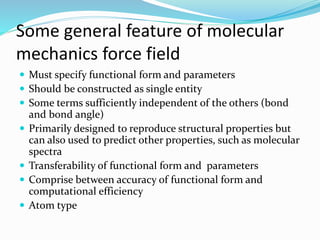
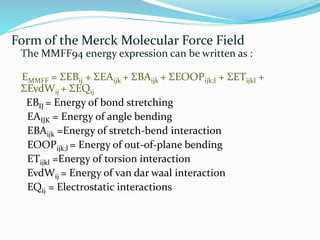
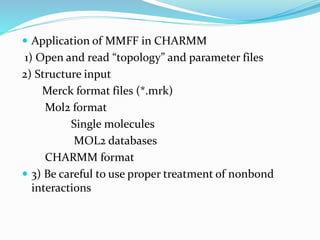
This document discusses molecular mechanics force fields, specifically the Merck Molecular Force Field (MMFF). It provides details on the functional form and parameters of MMFF, including that it is a Class II force field designed to accurately model conformational energies and non-bonded interactions of pharmaceutical compounds. The total energy expression for MMFF is provided, including terms for internal interactions like bonds, angles, and torsions, as well as nonbonded van der Waals and electrostatic terms. Application of MMFF in the CHARMM program is also described.