This document provides a summary of a presentation on using MongoDB and big data technologies to accelerate pharmaceutical research and development at AstraZeneca. The presentation discusses:
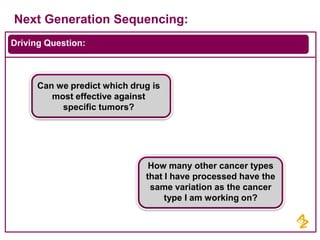
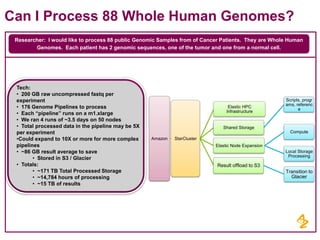
- AstraZeneca's focus on using next generation sequencing and big data to predict drug effectiveness and identify new drug targets
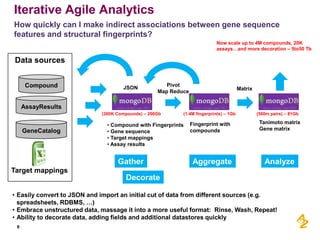
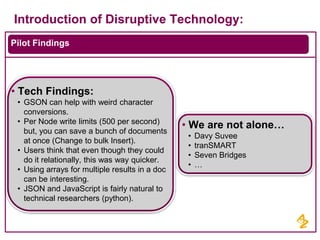
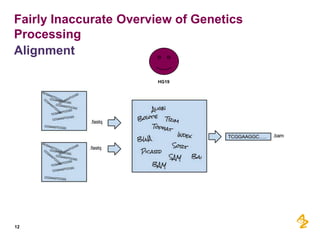
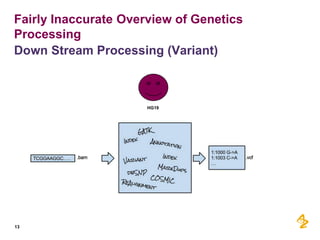
- Pilot projects using MongoDB to store and query unstructured genomic data at scale, which proved the technology's ability to enable researchers more quickly
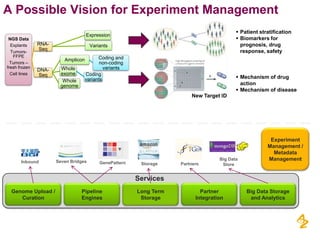
- A vision for an experiment management system to integrate various data sources and processing pipelines using big data technologies





![VCF as JSON
Header and Variant Information
{
{
"_id" : ObjectId("52617b613004b77f64efed62"),
"ALT" : [
"A"
],
"QUAL" : "29",
"NA00001" : "0|0:48:1:51,51",
"POS" : 14370,
"NA00002" : "1|0:48:8:51,51",
"FILTER" : "PASS",
"CHROM" : "20",
"NA00003" : "1/1:43:5:.,.",
"FORMAT" : "GT:GQ:DP:HQ",
"__vcfid" : "40770f6f-165a-4930-8092-05e98e4e0b27",
"ID" : "rs6054257",
"INFO" : {
"DP" : "14",
"AF" : "0.5",
"NS" : "3"
},
"REF" : "G"
}
18
"_id" : ObjectId("52617b613004b77f64efed67"),
"phasing" : "partial",
"fileformat" : "VCFv4.1",
"fileDate" : "20090805",
"source" : "myImputationProgramV3.1",
"FORMAT" : {
"Description" : ""Haplotype Quality"",
"Type" : "Integer",
"Number" : "2",
"ID" : "HQ"
},
"__vcfid" : "40770f6f-165a-4930-8092-05e98e4e0b27",
"contig" : {
"species" : ""Homo sapiens"",
"assembly" : "B36",
"md5" : "f126cdf8a6e0c7f379d618ff66beb2da",
"length" : "62435964",
"ID" : "20",
"taxonomy" : "x"
},
"INFO" : {
"Description" : ""HapMap2 membership"",
"Type" : "Flag",
"Number" : "0",
"ID" : "H2"
},
"reference" : "file:///seq/references/1000GenomesPilotNCBI36.fasta",
"FILTER" : {
"Description" : ""Less than 50% of samples have data"",
"ID" : "s50"
}
}](https://image.slidesharecdn.com/accelerate-20pharmaceutical-20r-26d-20with-20mongodb-131126132706-phpapp02/85/Accelerate-pharmaceutical-r-d-with-mongo-db-18-320.jpg)



