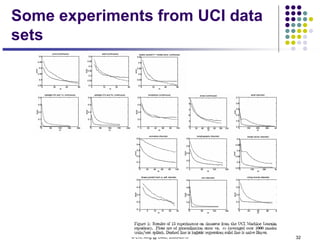
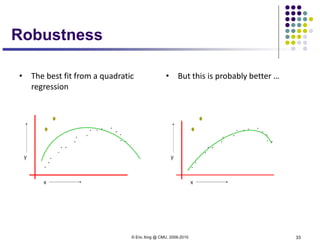
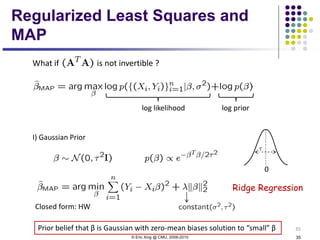
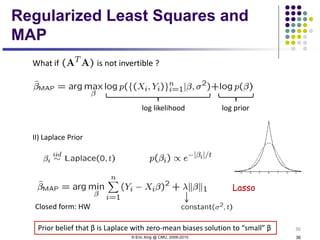
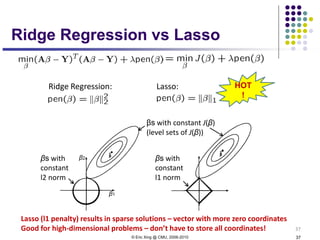
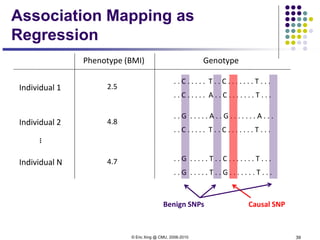
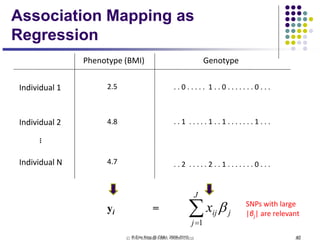
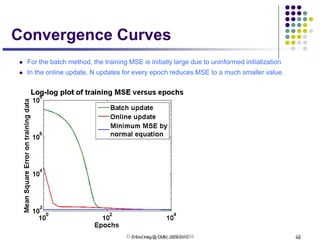
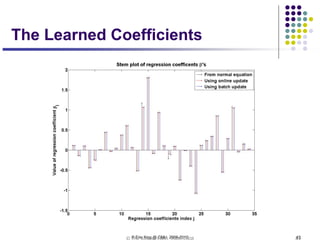
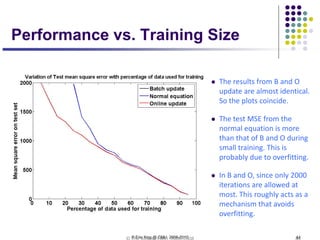
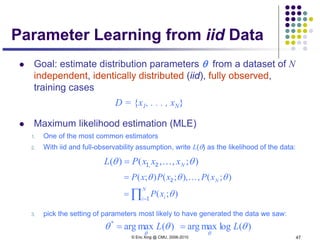
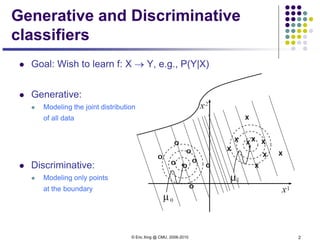
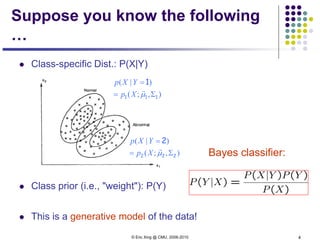
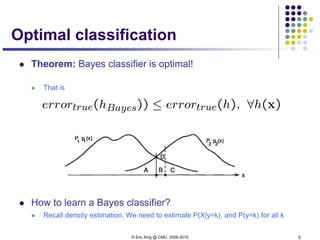
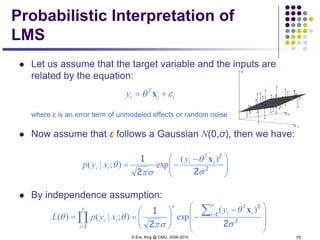
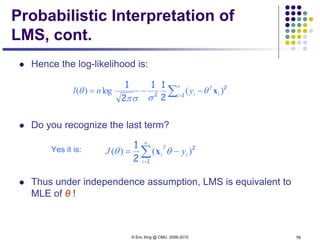
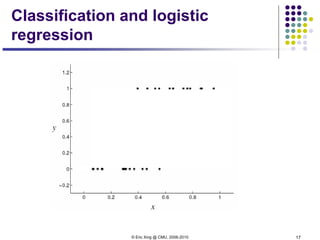
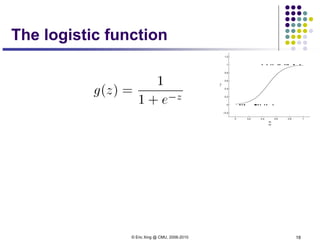
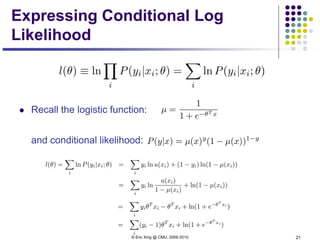
This document discusses generative and discriminative classifiers. Generative classifiers model the joint distribution of data and labels, while discriminative classifiers directly model the conditional probability of labels given data. Naive Bayes is an example of a generative classifier, while logistic regression is a discriminative classifier that directly models the probability of a label given input features. The document provides mathematical details on naive Bayes, logistic regression, and how logistic regression can be trained to maximize conditional likelihood through gradient descent.


![© Eric Xing @ CMU, 2006-2010 9
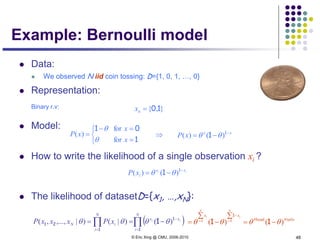
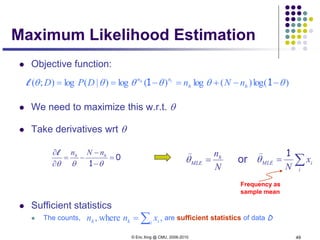
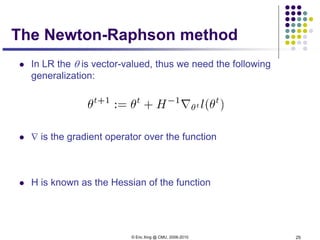
The predictive distribution
Understanding the predictive distribution
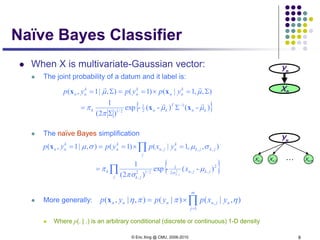
Under naïve Bayes assumption:
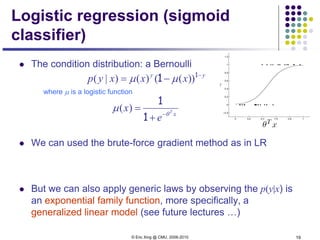
For two class (i.e., K=2), and when the two classes haves the same
variance, ** turns out to be a logistic function
*
),|,(
),|,(
),|(
),,|,(
),,,|(
' '''∑ Σ
Σ
=
Σ
Σ=
=Σ=
k kknk
kknk
n
n
k
n
n
k
n
xN
xN
xp
xyp
xyp
µπ
µπ
µ
πµ
πµ
1
1
( ){ }1
1
222
122
1
1
2
222
1
2
12
21
1
21
1
1
1
1
1
π
π
σσσµπ
σµπ
µµµµ
σ
σ
)(2
log)-(exp
log)-(exp
log)][-]([)-(exp −
−−−
−−−
++−+
=
∑
∑
+
=
∑j
jjjjj
nCx
Cx
jj
j j
jj
n
j
j j
jj
n
j
x
n
T
x
e θ−
+
=
1
1
)|( nn xyp 11
=
**
log)(exp
log)(exp
),,,|(
' ,''
,'
'
,
,
∑ ∑
∑
−−−−
−−−−
=Σ=
k j jk
j
k
j
n
jk
k
j jk
j
k
j
n
jk
k
n
k
n
Cx
Cx
xyp
σµ
σ
π
σµ
σ
π
πµ
2
2
2
2
2
1
2
1
1
](https://image.slidesharecdn.com/lecture2xing-150527174555-lva1-app6891/85/Lecture2-xing-9-320.jpg)









![© Eric Xing @ CMU, 2006-2010 31
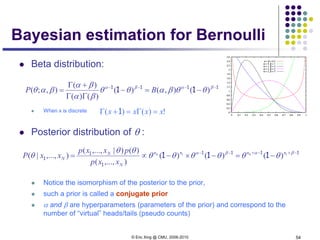
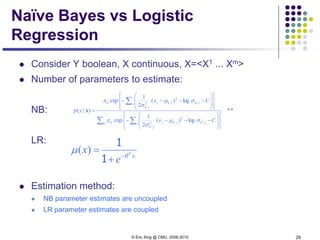
Naïve Bayes vs Logistic
Regression
Non-asymptotic analysis (see [Ng & Jordan, 2002] )
convergence rate of parameter estimates – how many training
examples needed to assure good estimates?
NB order log m (where m = # of attributes in X)
LR order m
NB converges more quickly to its (perhaps less helpful)
asymptotic estimates](https://image.slidesharecdn.com/lecture2xing-150527174555-lva1-app6891/85/Lecture2-xing-31-320.jpg)