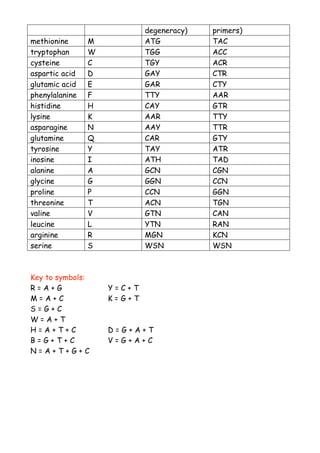
This document provides guidance on designing degenerate primers that can amplify similar genes from multiple species. It recommends selecting a conserved 6-7 amino acid peptide sequence and designing a 20 nucleotide oligo to match. The oligo should accommodate all possible codons for conserved residues among the target proteins. Amino acids with many codons like leucine and arginine should be avoided in favor of those with few codons. Inosine can be used where complete degeneracy exists to reduce the number of primers needed. Degeneracy should also be avoided at the 3' end.